Figures & data
Table 1. Demographics of human cases used in this study.
Figure 1. Immunoblot analysis of total tau and sarkosyl-insoluble tau in human brains. (a) Anti-tau antibodies used in this study were described with their recognition sites within the full-length 2N4R isoform of tau. (b) The brain lysate or sarkosyl-insoluble fraction from three cases of AD and three cases of non-demented controls were investigated by Western blot analysis with the use of phosphorylation-independent anti-tau antibodies Ser262 or 2B11. (c) The brain lysate or sarkosyl-insoluble fraction from the brains described in (b) were investigated by Western blot analysis with the use of phosphorylation-dependent anti-tau antibodies AT8 or AT100 or pS396. Molecular mass markers in kilodaltons are shown on the left.
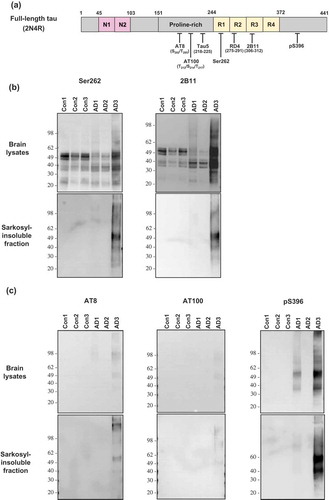
Figure 2. Expression of tau RD(LM) fused to eGFP in HEK293T cells. Repeat domain (RD) of 4R tau mutated at two positions (P301L and V337M; tau RD[LM]) were fused to eGFP and expressed in HEK293T cells. A monoclonal HEK293T cell line stably expressing the mutant tau RD was selected and employed in this study. (a) A representative image of a monoclonal HEK293T cell line expressing tau RD(LM)-eGFP. For comparison, an image of HEK293T cells were shown in parallel. Scale bar = 50 µm. (b) The lysate of the monoclonal HEK293T cells carrying tau RD(LM)-eGFP was examined by Western blot analysis in comparison with that of normal HEK293T cells, with GAPDH as a loading control. The blots were probed using anti-tau antibody (RD4) or anti-eGFP antibody. Molecular mass markers in kilodaltons are shown on the left.
![Figure 2. Expression of tau RD(LM) fused to eGFP in HEK293T cells. Repeat domain (RD) of 4R tau mutated at two positions (P301L and V337M; tau RD[LM]) were fused to eGFP and expressed in HEK293T cells. A monoclonal HEK293T cell line stably expressing the mutant tau RD was selected and employed in this study. (a) A representative image of a monoclonal HEK293T cell line expressing tau RD(LM)-eGFP. For comparison, an image of HEK293T cells were shown in parallel. Scale bar = 50 µm. (b) The lysate of the monoclonal HEK293T cells carrying tau RD(LM)-eGFP was examined by Western blot analysis in comparison with that of normal HEK293T cells, with GAPDH as a loading control. The blots were probed using anti-tau antibody (RD4) or anti-eGFP antibody. Molecular mass markers in kilodaltons are shown on the left.](/cms/asset/dc492c52-68d2-4bcc-8715-7bafa422d5d3/kprn_a_1545524_f0002_oc.jpg)
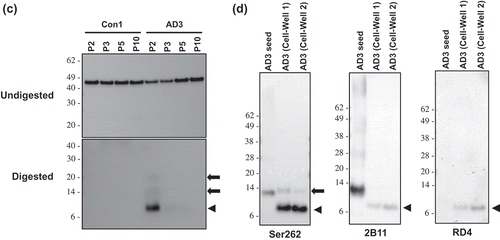
Figure 3. Identification of protease-resistant fragment of tau RD(LM) aggregates formed in HEK293T cells by seeded aggregation. The HEK293T cells expressing tau RD(LM) were exposed to the sarkosyl-insoluble fraction of brain samples for ~20 hours and passaged up to 20 times. (a) Cells at passage 2 were fixed in the condition to remove soluble proteins and the formation of tau RD(LM) aggregates were analysed by fluorescent microscopy. Scale bar = 100 µm. (b) Cells at multiple passages were fixed as in (a) and the formation of tau RD(LM) aggregates were examined by confocal microscopy. Scale bar = 50 µm. (c) Cells were recovered at different passages and lysed. Following the adjustment of total protein to 4 mg/mL, the lysates left undigested or digested with pronase at 50 µg/ml and examined by Western blot analysis using anti-tau antibody 2B11. Arrowheads indicate bands representing a monomeric form of pronase-resistant tau fragment. Arrows indicate bands representing a dimeric or trimeric form of proteolytic tau fragment. Molecular mass markers in kilodaltons are shown on the left. (d) The sarkosyl-insoluble fraction of AD3 or AD3-exposed cells recovered at passage 2 were digested with pronase at 50 µg/ml and then investigated by Western blot analysis using three anti-tau antibodies Ser262, 2B11 or RD4. Arrowheads indicate bands representing a monomeric form of pronase-resistant tau fragment. Arrows indicate bands representing a dimeric form of proteolytic tau fragment. Molecular mass markers in kilodaltons are shown on the left.
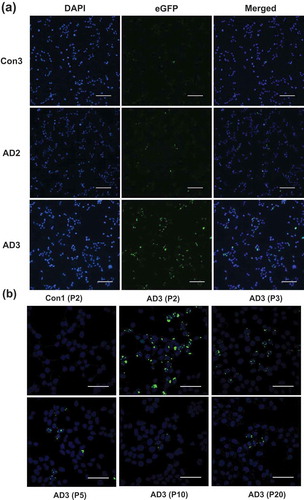
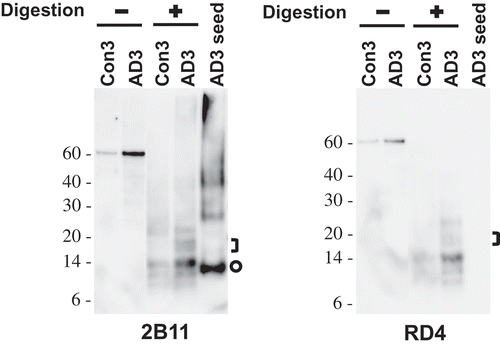
Figure 4. Cell-free conversion of recombinant tau protein into the pronase-resistant form by seeded reaction. Recombinant tau proteins were mixed with the sarkosyl-insoluble fraction of brain samples and then incubated at 37°C for 72 hours with intermittent shaking (1 minute shaking at 400 rpm and 1 minute rest). The reaction products left undigested or digested with pronase at 10 µg/mL and examined by Western blot analysis using anti-tau antibodies 2B11 or RD4. The loaded amount of undigested sample was 1/5th of those digested. Pronase-digested AD3 seed was also run on the same gel for the comparison of gel mobility between seeds and in vitro (cell-free) generated species. Brackets designate bands generated in the reactions specifically seeded with AD3. Circles indicate bands representing a monomeric form of pronase-resistant tau fragment of the AD3 seed. Molecular mass markers in kilodaltons are shown on the left.