Figures & data
Table 1. PRNP haplotypes tabulated from 1,433 white-tailed deer tissues collected from 75 counties in Arkansas (2016–2019). Haplotypes (Hap) are identified by letter (A-V) following Brandt et al. (2015, 2018). Haplotypes denoted as AR1-4 are previously unreported. Mutations that differ from haplotype A are shaded, with green indicating synonymous substitutions (no amino acid change) and blue as non-synonymous (amino acid changed in protein; NSS). Also listed are amino acid position and nucleotide site (Brandt et al. 2015, 2018). Polymorphisms not identified in prior studies are denoted as *
Figure 1. Sampling locations for white-tailed deer tissues from Arkansas evaluated in this study. The red shaded area indicates the 16 counties included in the 2019 Chronic Wasting Disease Management Zone (CWDMZ), with a yellow boundary surrounding a focal area encompassing Newton County. Black dots represent collection localities for each individual tissue. Note that the boundaries of the CWDMZ have since expanded
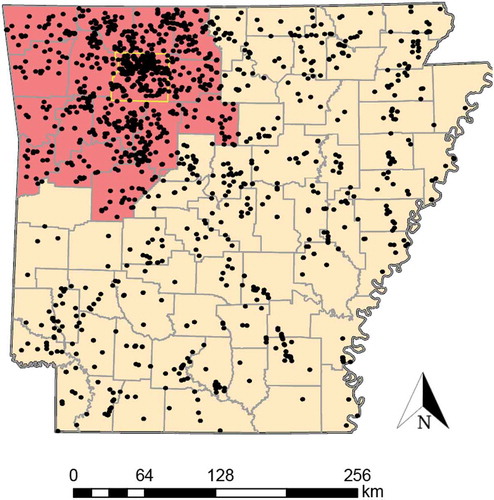
Table 2. PRNP haplotype frequencies and odds-ratios, as associated with CWD status of white-tailed deer in Arkansas, 2016–2019. Haplotypes were derived from unphased sequences representing 720 nucleotides of the PRNP gene. Haplotypes (Hap) are identified by letter (A-V) following Brandt et al. (2015, 2018). Haplotypes denoted AR1-4 are previously unreported. Listed are total numbers (N) and relative frequency (%) and associated values for deer that were CWD-negative (-), CWD-positive (+) or untested (?). Odds Ratio (OR) reflects the relative representation of a haplotype in CWD-positive deer; OR>1 indicated over-representation, OR<1 under-representation. SE = standard error, CI = 95% confidence interval, Z = OR Z-score and p= OR p-value. Values in bold are significant
Figure 2. Haplotype network showing relationship of prion gene variants (PRNP) detected across 1,433 white-tailed deer collected from 75 counties in Arkansas (2016–2019). Data are based on sequence analysis of 720 nucleotides. Circles represent 20 haplotypes (=alleles) with size reflecting frequency of occurrence in entire data set (), and tick marks representing number of mutations (=nucleotide substitutions) distinguishing one from another (). Colour codes reflect relative frequency among CWD-positive (red) and CWD-negative/undetected (green). Letters correspond to haplotype names (per Brandt et al. 2015), with haplotypes unique to Arkansas indicated with numbers (AR#). Haplotypes sharing the 96S mutation are indicated with (*)
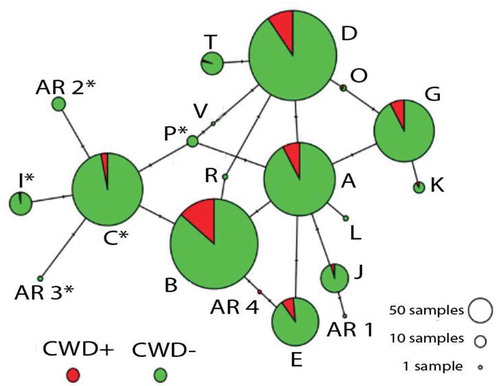
Figure 3. Stacked histogram of 2,866 PRNP haplotypes detected in white-tailed deer collected in Arkansas, 2016–2019. Haplotypes were determined by phasing individual genotypes derived from sequencing 1,433 deer across 720 bp of the PRNP gene. Letters (A through V) refer to haplotypes identified in Brandt et al. (2015), whereas numbers (1–4) are haplotypes unique to Arkansas, and thus previously unreported. Frequencies are plotted for all 1,433 samples (=statewide) as well as for a subset of 314 samples from Newton County (N = 628 chromosomes). Colour codes reflect frequency among CWD-positive (CWD+) and CWD-negative (CWD-) samples; unknown indicates samples that were not tested for CWD. For each haplotype, paired bars report values statewide (left) and for the Newton County subset (right)
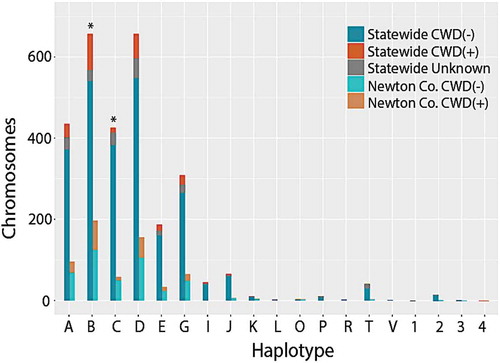
Figure 4. Topographies that represent interpolated haplotype frequencies for the PRNP gene in Arkansas. Frequency is depicted by colour, with blue reflecting low occurrence (0–5%) whereas red indicating 46–50+% of haplotypes were of this type
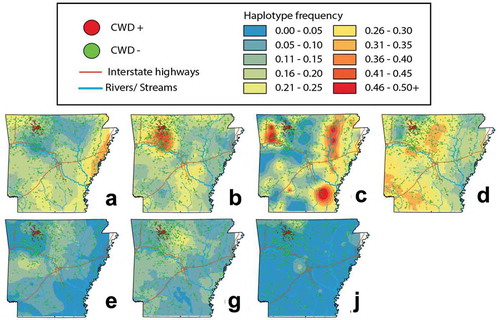
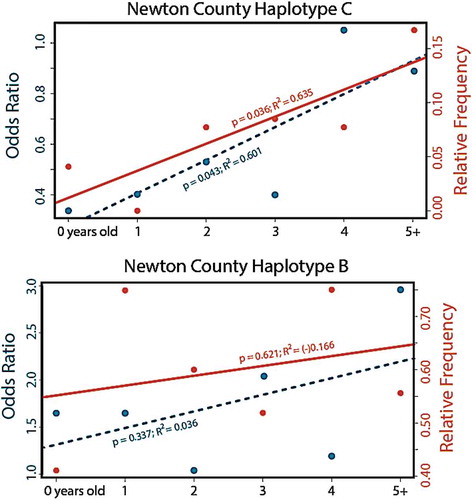
Figure 5. Relative frequency and odds ratio for two haplotypes of the prion gene PRNP haplotypes detected in white-tailed deer age cohorts (<1 year to 5+ years) sampled in Arkansas from 2016–2019. Prion gene variant Haplotype C (top panel) has been associated with reduced susceptibility to CWD, whereas Haplotype B (lower panel) has been associated with higher susceptibility (Brandt et al. 2018). Data are based on phased haplotypes derived from 720 nucleotides of the PRNP gene sequenced across 1,433 deer