Figures & data
Figure 1. Reported county-level distribution of free-ranging axis deer (Axis axis) in Texas (a) and islands where free-range axis deer occur in Hawaii (b). The Texas map includes locations of the chronic wasting disease (CWD) surveillance zones established by Texas Parks and Wildlife Department (TPWD) in response to CWD having been detected in free-ranging and captive cervids
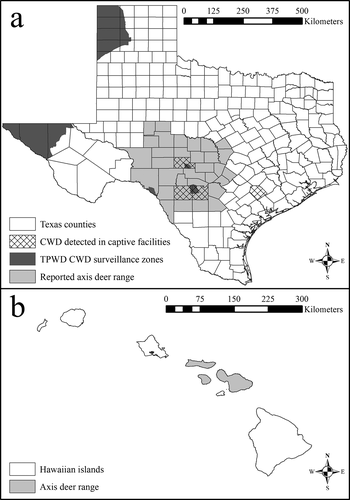
Figure 2. Spatial distribution of the free-ranging axis deer (Axis axis) sequenced for PRNP exon 3 for this study and the reported ranges of free-ranging axis deer in Texas (a) and Hawaii (b)
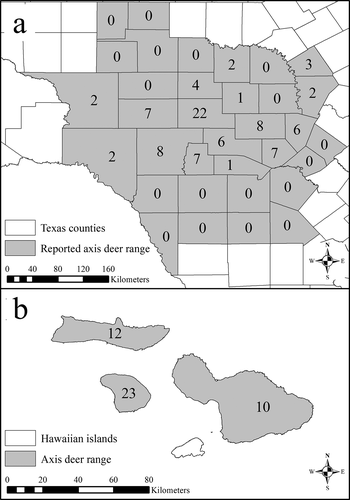
Figure 3. Phylogenetic tree generated from prion protein gene (PRNP) sequences from Cervidae with Bayesian inference analyses (MrBayes 3.2) [Citation37]. Nodal support (posterior probability values ≥ 0.95) [Citation38] is indicated by an asterisk above the node. Clades are identified by letters below the node that contains the identified clade. Although there was a lack of continued support beyond the basal node, 8 groups composed of individual species or closely related species in the larger Cervidae phylogeny [Citation24] were distinguishable from the analyses
![Figure 3. Phylogenetic tree generated from prion protein gene (PRNP) sequences from Cervidae with Bayesian inference analyses (MrBayes 3.2) [Citation37]. Nodal support (posterior probability values ≥ 0.95) [Citation38] is indicated by an asterisk above the node. Clades are identified by letters below the node that contains the identified clade. Although there was a lack of continued support beyond the basal node, 8 groups composed of individual species or closely related species in the larger Cervidae phylogeny [Citation24] were distinguishable from the analyses](/cms/asset/774fa04c-402d-43a2-a217-a7b4e28647a5/kprn_a_1910177_f0003_b.gif)
Figure 4. Polymorphic amino acid residues that have been linked to Chronic Wasting Disease susceptibility and the Cervid PrPC variant differentiating amino acid residue (residue 226) in white–tailed deer (Odocoileus virginianus), mule deer (O. hemionus), and North American Elk (Cervus canadensis) [Citation1] in comparison to the axis deer (Axis axis) amino acid sequence identified in this study. The substitutions shown convey some level of reduced susceptibility in the form of longer incubation times and reduced prevalence in the proportion of each respective species that possess the substitution genotype compared to the proportion that possess the wild type genotype [Citation1,Citation9–11]. Axis deer possess the wild type genotype at all the polymorphic sites listed for the other species, the elk PrPC variant, and a previously unidentified substitution at residue 249. The shaded areas depict the terminal sequences removed during post-translational editing of PrPC [Citation27]
![Figure 4. Polymorphic amino acid residues that have been linked to Chronic Wasting Disease susceptibility and the Cervid PrPC variant differentiating amino acid residue (residue 226) in white–tailed deer (Odocoileus virginianus), mule deer (O. hemionus), and North American Elk (Cervus canadensis) [Citation1] in comparison to the axis deer (Axis axis) amino acid sequence identified in this study. The substitutions shown convey some level of reduced susceptibility in the form of longer incubation times and reduced prevalence in the proportion of each respective species that possess the substitution genotype compared to the proportion that possess the wild type genotype [Citation1,Citation9–11]. Axis deer possess the wild type genotype at all the polymorphic sites listed for the other species, the elk PrPC variant, and a previously unidentified substitution at residue 249. The shaded areas depict the terminal sequences removed during post-translational editing of PrPC [Citation27]](/cms/asset/ab162177-ea26-4c85-bb43-f496e97f6a9c/kprn_a_1910177_f0004_b.gif)
