Figures & data
Table 1. Genotype and allele frequencies of 19 PRNP polymorphism in Nigerian sheep.
Table 2. Linkage Disequilibrium (LD) of nineteen (19) PRNP polymorphisms in Nigerian sheep.
Table 3. Haplotype frequencies of fourteen (14) non-synonymous single nucleotide polymorphism of PRNP gene in Nigerian sheep.
Figure 1. Comparisons of the allele frequencies of PRNP codons 154 and 240 in Central and Southern Italy, Chinese and Nigerian sheep. (a) Comparisons of the allele frequency of the PRNP codon 154 between Central and Southern Italy, Chinese sheep and Nigerian sheep [Citation30,Citation63] (b) Comparisons of the allele frequency of PRNP codon 240 between Chinese and Nigerian sheep [Citation30].
![Figure 1. Comparisons of the allele frequencies of PRNP codons 154 and 240 in Central and Southern Italy, Chinese and Nigerian sheep. (a) Comparisons of the allele frequency of the PRNP codon 154 between Central and Southern Italy, Chinese sheep and Nigerian sheep [Citation30,Citation63] (b) Comparisons of the allele frequency of PRNP codon 240 between Chinese and Nigerian sheep [Citation30].](/cms/asset/ca24d4e0-cdb2-46d2-82c2-ed88680e0c9a/kprn_a_2186767_f0001_oc.jpg)
Table 4. Distributions of haplotype and genotype frequencies at PRNP codons 143, 154 and 171 between scrapie affected and healthy sheep.
Figure 2. Prediction of amyloid propensity of sheep prion protein according to nonsynonymous SNPs. AMYCO predicted amyloid propensity as values from 0.0 to 1.0. The AMYCO scores<0.45 and>0.78 indicated low and high aggregation propensities of the protein, respectively. ‘HYNN’ indicates haplotype of serine allele at the codon 139, serine allele at the codon 146, histidine allele at the codon 154, and Isoleucine allele at the codon 193. ‘HYKK’ indicates haplotype of Arginine allele at the codon 139, asparagine allele at the codon 146, arginine allele at the codon 154, and threonine allele at the codon 193. ‘HDKK’ indicates haplotype of arginine allele at the codon 139, asparagine allele at the codon 146, histidine allele at the codon 154, and threonine allele at the codon 193. ‘RYNN’ indicates haplotype of arginine allele at the codon 139, asparagine allele at the codon 146, arginine allele at the codon 154, and isoleucine allele at the codon 193.
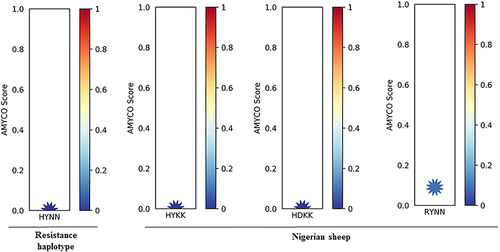
Table 5. Measurement of the effect of amino-acid substitutions of PRNP nonsynonymous SNPs in Nigerian sheep.
Supplemental Material
Download Zip (3.4 MB)Data availability statement
Accession no. MZ463500 – MZ463625 https://www.ncbi.nlm.nih.gov/
