Figures & data
Figure 1. Organs and tissues affected by systemic and localized amyloidosis are in many instances overlapping with those afflicted by acute COVID-19 and PASC.
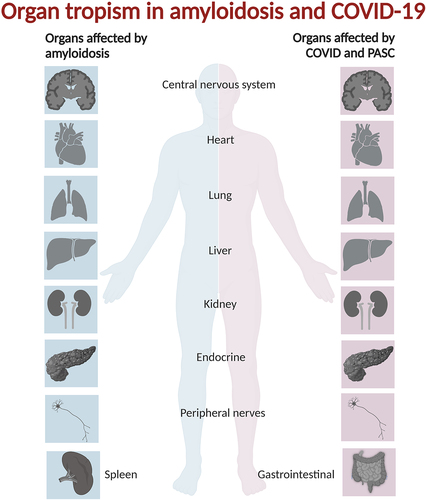
Table 1. Human amyloid fibril proteins and their precursors. Table is modified from [Citation29]. Proteins are listed in groups as full-length protein in fibril, fragments in fibril, and endoproteolytic products from precursor proteins.
Figure 2. Endoproteolysis is a common and often required pathway for amyloid formation of natively folded and functional proteins.
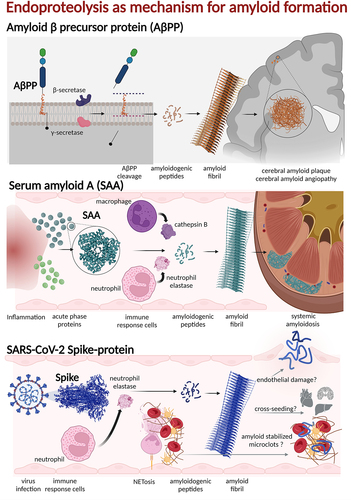
Figure 3. TEM micrographs of (on top) full length Spike-protein (Wuhan strain) after co-incubation with neutrophil elastase at 37°C for 24 h. The in silico predicted peptide 192–211 of SARS-CoV-2 Spike-protein, incubated at 37°C for 24 h. The resulting fibrils are straight, twisted and rod-like [Citation81].
![Figure 3. TEM micrographs of (on top) full length Spike-protein (Wuhan strain) after co-incubation with neutrophil elastase at 37°C for 24 h. The in silico predicted peptide 192–211 of SARS-CoV-2 Spike-protein, incubated at 37°C for 24 h. The resulting fibrils are straight, twisted and rod-like [Citation81].](/cms/asset/1faa6db9-4da5-4244-a1b2-2b513467cf46/kprn_a_2194212_f0003_oc.jpg)
Figure 4. There are several mechanisms by which amyloids and viruses co-operate.
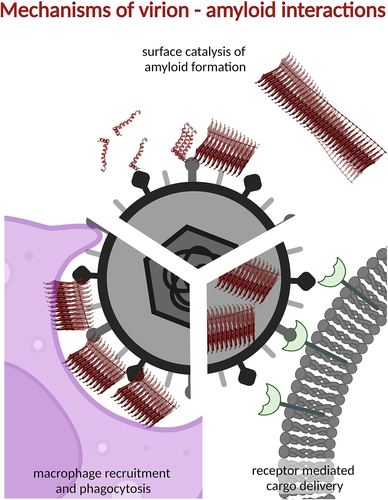
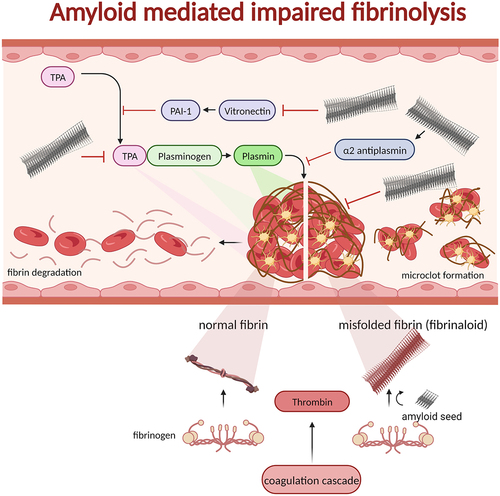
Figure 5. Blood clots formed by thrombin processing of fibrinogen to fibrin recruiting blood platelets in a polymerizing fibrous web. The process of fibrinolysis is regulated by several proteins ultimately affording active plasmin as the proteolytic enzyme to digest insoluble fibrin and, hence resolving the clot. The normal process is shown on the left side of the clot in the figure. If fibrin on the other hand is generated from fibrinogen by thrombin (possibly even without thrombin) in the presence of amyloid seeds the fibrin can form misfolded fibrin (fibrinaloid) which cannot be digested by plasmin (right side of the figure). In addition to seeding misfolding of fibrin, amyloid fibrils are inactivating and activating proteins involved in fibrinolysis: tPA, vitronectin, α2-antiplasmin, and plasmin. Hence, there are many associations between fibrinolysis and amyloid disease as discussed in the main text.