Figures & data
Figure 1. Cell spreading on elongated adhesive patterns. (A) Side and (B) top views of a cell spreading on a rectangular fibronectin micropattern of 1:10 aspect ratio. (C) Side and (D) top views of an immunostained endothelial cell spread on a rectangular micropattern (1:10 aspect ratio) deposited on a stiff (E = 3MPa) PDMS substrate. The actin cytoskeleton (phalloidin) is labeled in green, focal adhesions (vinculin) in red and the nucleus (DAPI) in blue. Scale bars are 10 µm. (E) Temporal evolution of the normalized cell spreading area on rectangular adhesive micropatterns (aspect ratio 1:10) deposited on 5 kPa (in blue, n = 9), 9 kPa (in red, n = 8) and 3 MPa (in black, n = 11) culture substrates. (F) Mean characteristic spreading times, ts, on 5kPa (in blue, n = 9), 9kPa (in red, n = 8) and 3 MPa (in black, n = 11) substrates. Data are mean ± SD, *p ≤ 0 .05, ** p ≤ 0 .01 and *** p ≤ 0 .001.
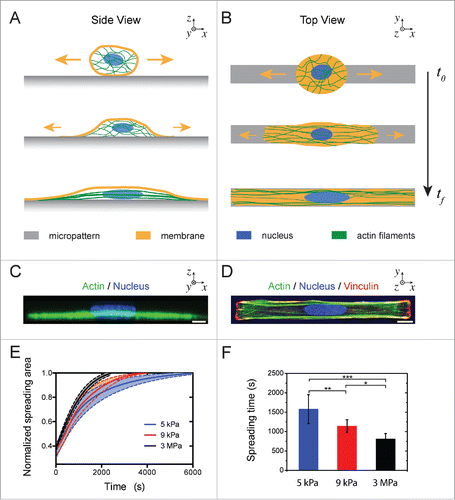
Figure 2. Cell relaxation dynamics. (A) Time-lapse sequence in DIC mode of the cell relaxation process after detachment with Accutase (t = 0) of an endothelial cell spread on an elongated micropattern (1:10 aspect ratio, depicted in white) deposited on a stiff (E = 3 MPa) substrate. The scale bar is 10 µm. (B) Evolution of the normalized cell deformation as a function of time after initiation of the detachment from an elongated pattern on a stiff substrate. The red curve corresponds to a sigmoidal fit with Eq 4. DIC kymographs of the relaxation of an endothelial cells detached from a (C) 5 kPa and a (D) 3 MPa micropatterned substrates. The slopes on kymographs represent cell relaxation velocity after detachment on 5 kPa (blue line) and 3 MPa (black line), respectively. (E) Evolution of the characteristic cell relaxation time as a function of the matrix stiffness (n = 13 for 5 kPa; n = 18 for 9 kPa; and n =15 for 3MPa). Data are expressed as mean ± SD, *p ≤ 0 .05, ** p ≤ 0 .01 and n.s. non significant.
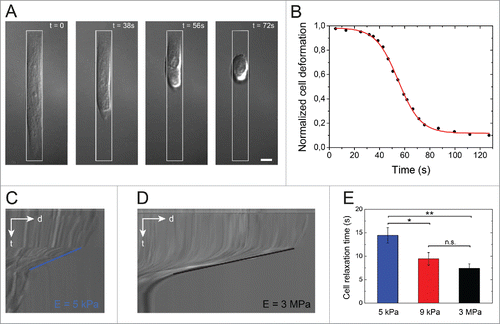
Figure 3. Role of the actomyosin cytoskeleton on cell relaxation dynamics. (A) Cellular relaxation times (mean ± SD) for control endothelial cells (black bar, n = 18), Latrunculin A-treated cells (white bar, n = 14) and Y-27632-treated cells (dashed bar, n=15) deposited on 3 MPa micropatterned substrates. Data are expressed as mean ± SD and ***p ≤ 0 .001. (B) Typical fluorescent images of the spatial distribution of the actin cytoskeleton (in green), the nucleus (in blue) and the myosin (in white) of endothelial cells plated on 1200 µm2 and 1:10 aspect ratio rectangular micropatterns deposited on 3MPa substrates. Scale bars are 10 µm. (C) Semi-logarithmic evolution of the cell elastic modulus of endothelial cells spread on elongated patterns deposited on substrates with increasing stiffnesses (5 kPa, 9 kPa, 45 kPa, 110 kPa and 3 MPa). (D) Evolution of the characteristic relaxation time as a function of the cell elastic modulus.
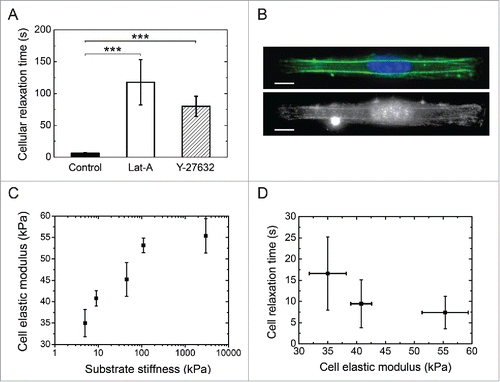
Figure 4. Evolution of the nuclear deformation during cellular spreading and relaxation processes. (A) Image sequence of the cellular (DIC) and the nuclear (DAPI) deformations during the cell spreading stage on an elongated micropattern (depicted in red). A schematic representation of the cell for each step is indicated for clarity. (B) DIC and DAPI images of a relaxed cell and its nucleus, respectively, after cell detachment with Accutase. (C) Color-coded fluorescent images (H2B-GFP labeling) of the nucleus of an endothelial cell spreading on a 3MPa micropatterned substrate. The scale bar represents 10 µm. (D) Color-coded fluorescent images (H2B-GFP labeling) of the nucleus of an endothelial cell relaxing from a 3MPa micropatterned substrate after detachment with Accutase. The scale bar represents 10 µm. (E) Typical evolution of cellular (in black) and nuclear (in blue) deformations during spreading and relaxation stages on a 3MPa micropatterned substrate. (F) Schematic representation of the reorganization of chromatin during the slow (∼min.) nuclear deformation associated to cell spreading and the fast (∼s) nuclear relaxation after cell detachment.
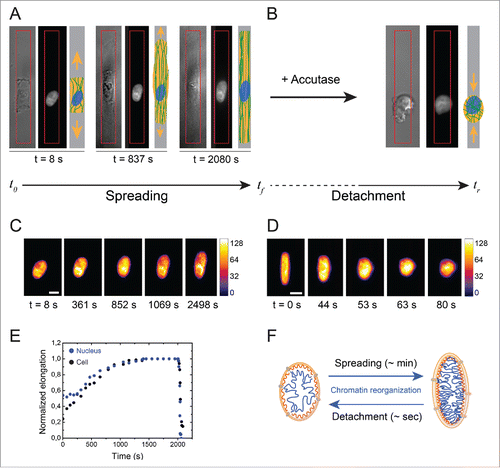
Figure 5. Dynamics of nuclear relaxation. Evolution of the characteristic nuclear relaxation time as a function of (A) the matrix stiffness (E = 5 kPa n = 5, 9 kPa n = 6 and 3 MPa n = 7) and (B) a Y-27632 treatment that affects the actomyosin contractility (control cells: n = 7 and Y-27632 treated cells: n = 5). Data are expressed as mean ± SD, **p < 0.01 and n.s. non significant.
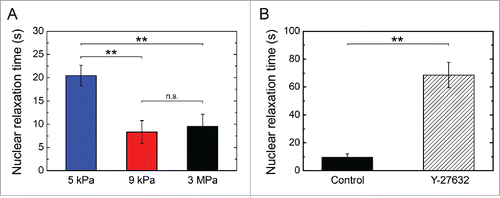
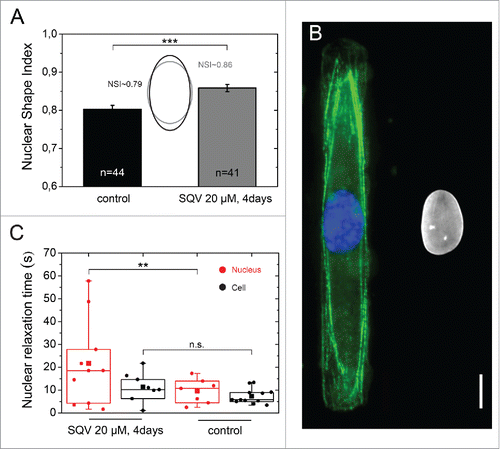
Figure 6. Role of the nuclear lamina on nuclear relaxation dynamics. (A) Characterization of the nuclear shape of control (in black, n = 44) and saquinavir-treated cells (in gray, n = 41) plated on a 3 MPa micropatterned substrate. Mean ± SD and ***p ≤ 0 .001. (B) Immunostaining images of the actin cytoskeleton (in green), the nucleus (in blue) and the lamin A/C (in white) of rectangular micropatterned endothelial cells treated for 3 d with the protease inhibitor saquinavir after plating and then treated with fresh saquinavir for 24h before fixation and immunostaining. Scale bar is 10 µm. (C) Characteristic nuclear (n = 11, in red) and cellular (n = 11, in black) relaxation time of endothelial cells grown on rectangular micropatterns and treated for 4 d with the protease inhibitor saquinavir (20 µM). ** p ≤ 0 .01 and n.s. non significant.