Figures & data
Figure 1. Effect of IFNG on the invasion of HTR-8/SVneo cells. The effect of varying concentrations (0, 1, 10, 50 ng/mL) of IFNG on the invasion of HTR-8/SVneo cells was determined by Matrigel matrix invasion assay as described in Materials and Methods. Panel A shows the fold change in invasion after treatment with IFNG for 24 h as compared to untreated HTR-8/SVneo cells. The results are shown as mean ± S.E.M. of three independent experiments. Panel B shows representative images of invaded HTR-8/SVneo cells with and without treatment of IFNG (10 ng/mL) for 24 h. p ≤ 0.05 was considered statistically significant.
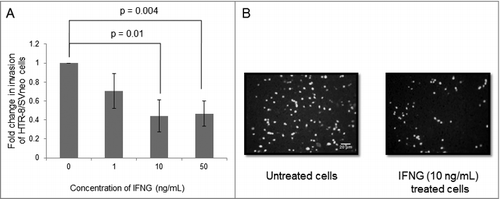
Figure 2. Effect of IFNG on phosphorylation of STAT1 in HTR-8/SVneo cells. HTR-8/SVneo cells (0.1 × 106) were treated with IFNG (10 ng/mL) for 0, 10, 30, 60 min. After treatment, cell lysates were prepared and proteins resolved by 0.1% SDS-10% PAGE and processed for analysis of phosphorylated forms of STAT1 and total STAT1 by Western blot as described in Materials and Methods. Panels A and B show the densitometric plots of p-STAT1 (tyr701) and p-STAT1 (ser727) normalized with respect to total STAT1. Panel C shows total STAT1 normalized with GAPDH. Representative blots of p-STAT1 (tyr701), p-STAT1 (ser727), total STAT1 and GAPDH are appended as Panel D. The data is expressed as fold change with respect to 0 h control and values are shown mean ± S.E.M. of at least three independent experiments.
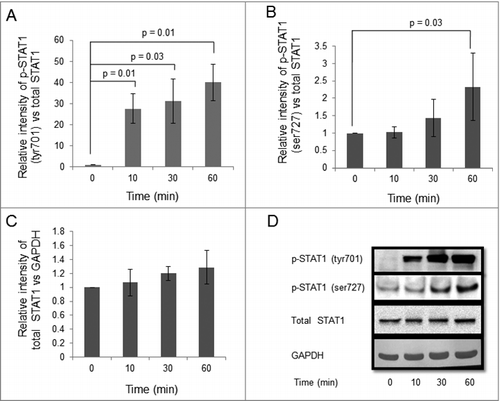
Figure 3. Role of STAT1 in IFNG-mediated decrease in invasion of HTR-8/SVneo cells. To study the importance of STAT1 in the invasion of HTR-8/SVneo cells after treatment with IFNG, silencing of STAT1 was done using siRNA as described in Materials and Methods. Silencing of STAT1 was confirmed by qRT-PCR and Western blotting. Panels A and B show expression profile of STAT1 at the transcript and protein levels in control siRNA and STAT1 silenced cells, respectively, on treatment with and without IFNG (10 ng/mL) for 24 h. Each bar represents relative expression after normalization with 18S rRNA and GAPDH and values expressed as mean ± S.E.M. of three independent experiments. Representative blots showing expression profile of STAT1 siRNA transfected cells is appended as part of Panel B. Panel C shows fold change in the invasion of control siRNA and STAT1 siRNA transfected cells on treatment with and without IFNG for 24 h. The results are expressed as mean ± S.E.M. of fold change in the invasion with respect to control siRNA transfected cells without IFNG (10 ng/mL) treatment, observed in three independent experiments.
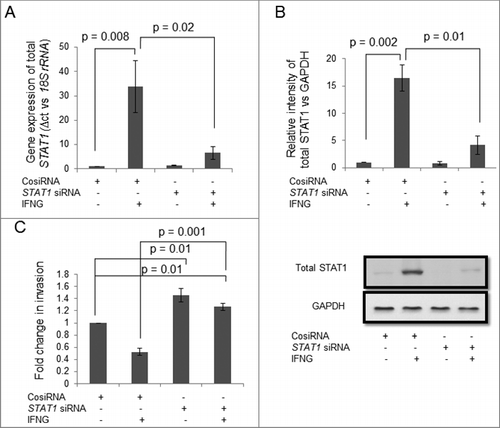
Figure 4. Expression profile of BATF2 in HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were treated with and without IFNG (10 ng/mL) for 24 h. Subsequently, RNA and cell lysates were prepared to determine BATF2 expression by qRT-PCR and Western blotting as described in Materials and Methods. Panel A shows the level of transcript encoding BATF2 using 18S rRNA as normalizing control. Panel B shows the expression profile of BATF2 at the protein level using GAPDH as a loading control in HTR-8/SVneo cells with and without treatment with IFNG. Values are expressed as mean ± S.E.M. of three independent experiments. Representative blots for BATF2 in untreated and treated HTR-8/SVneo cells with IFNG (10 ng/mL) for 24 h is also appended as part of panel B. p ≤ 0.05 was considered statistically significant as compared to untreated control.
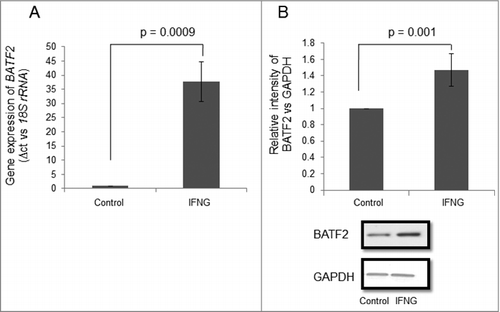
Figure 5. Effect of BATF2 silencing on the invasion of HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were transfected with control siRNA and BATF2 siRNA and subsequently used to study their invasion by transwell invasion assay as described in Materials and Methods. Silencing of BATF2 was confirmed by qRT-PCR and Western blotting. Panels A and B show transcript and protein levels of BATF2 in control siRNA and BATF2 silenced cells, respectively, on treatment with and without IFNG (10 ng/mL). Each bar represents relative expression after normalization with 18S rRNA or GAPDH, expressed as mean ± S.E.M. of three independent experiments. Panel C shows the fold change in the invasion of cells transfected with control or BATF2 siRNA respectively, subsequent to treatment with and without IFNG for 24 h. The results are expressed as mean ± S.E.M. of fold change in invasion, as compared to control siRNA transfected cells without treatment with IFNG, observed in three independent experiments.
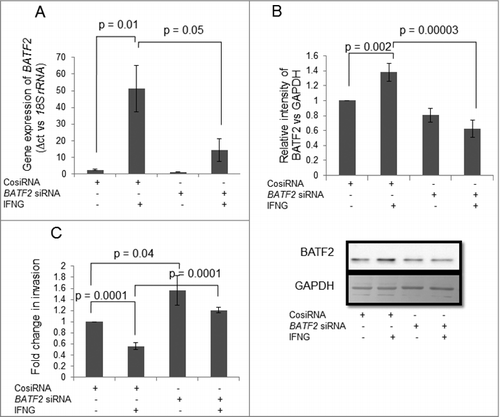
Figure 6. Effect of BATF2 silencing on the expression of STAT1 in HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were transfected with control siRNA and BATF2 siRNA and subsequently used to study the expression of total STAT1 in the presence or absence of treatment with IFNG (10 ng/mL) by qRT-PCR and Western blotting as described in Materials and Methods. Panels A and B show the transcript and the protein levels of STAT1 in control siRNA and BATF2 siRNA transfected HTR-8/SVneo cells, with and without IFNG treatment respectively. Each bar represents relative expression after normalization with 18S rRNA at transcript level and GAPDH at the protein level. Results are expressed as mean ± S.E.M. of three independent experiments. Representative blots of STAT1 expression in BATF2 silenced cells is appended in Panel B.
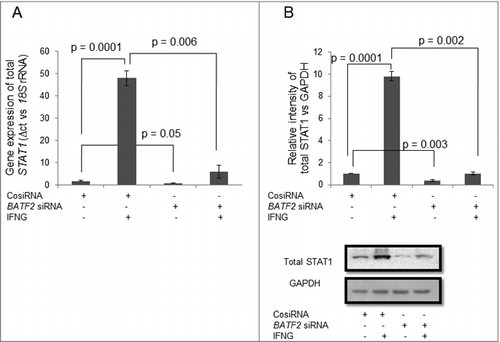
Figure 7. Expression of BATF2 in STAT1 silenced HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were transfected with control siRNA and STAT1 siRNA followed by treatment with or without IFNG (10 ng/mL) for 24 h and subsequently used to analyze expression of BATF2 both at the transcript and protein levels by qRT-PCR and Western blotting. Panel A shows the transcript level of BATF2 in control siRNA and STAT1 siRNA transfected cells with and without IFNG treatment. Panel B represents the densitometric analysis and representative blots of BATF2 expression in control siRNA and STAT1 siRNA transfected cells with and without IFNG treatment. Each bar represents relative expression after normalization with 18S rRNA at the transcript level and GAPDH at the protein level. Values are expressed as mean ± S.E.M. of three independent experiments.
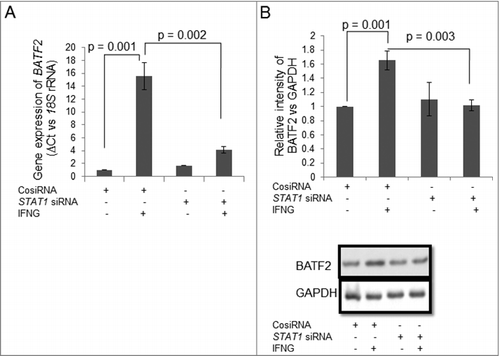
Figure 8. Expression of JUN in BATF2 silenced HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were transfected with control siRNA and BATF2 siRNA and subsequently used to study the expression of phosphorylated JUN ser 63 and JUN ser 73 in the presence or absence of IFNG (10 ng/mL) by Western blotting as described in Materials and Methods. Panel A, B and C show protein expression of p-JUN ser 63, p-JUN ser 73 and total JUN in control siRNA transfected and BATF2 silenced cells, respectively, on treatment with and without IFNG. Each bar represents relative expression after normalization with GAPDH with respect to untreated control siRNA transfected cells. Values are expressed as mean ± S.E.M. of three independent experiments. Panel D shows the representative blots of p-JUN ser 63, p-JUN ser 73, total JUN and GAPDH from one of the three experiments.
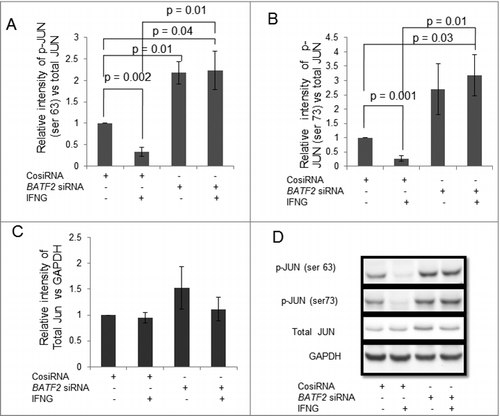
Figure 9. Expression of JUN in STAT1 silenced HTR-8/SVneo cells treated with IFNG. HTR-8/SVneo cells (0.1 × 106) were transfected with control siRNA and STAT1 siRNA and subsequently used to study the expression of phosphorylated JUN ser 63 and JUN ser 73 in the presence or absence of IFNG (10 ng/mL) by Western blotting. Panels A, B and C show protein expression of p-JUN ser 63, p-JUN ser 73, and total JUN in control siRNA transfected and STAT1 silenced cells, respectively, on treatment with or without IFNG. Each bar represents relative expression after normalization with total JUN and GAPDH respectively with respect to untreated control siRNA transfected cells. Values are expressed as mean ± S.E.M. of three independent experiments. Panel D shows the representative blots of p-JUN ser 63, p-JUN ser 73, total JUN and GAPDH from one of the three experiments.
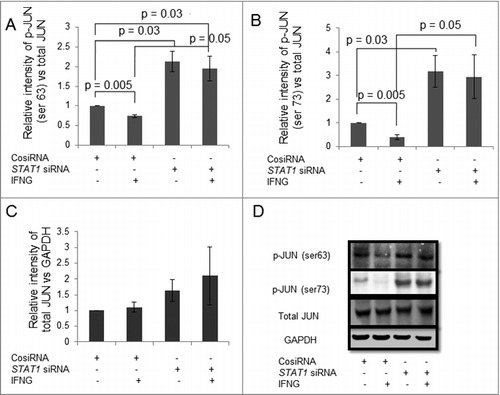
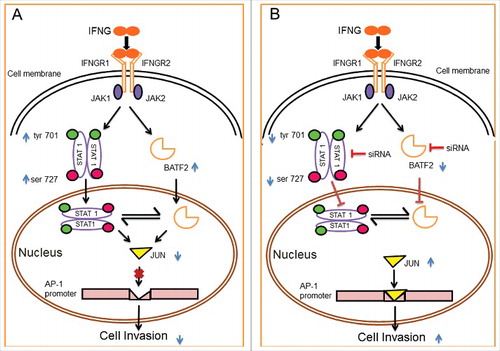
Figure 10. Schematic representation of the role of IFNG in regulation of HTR-8/SVneo cells invasion. Binding of IFNG to its receptors IFNGR1 and IFNGR2 lead to the phosphorylation of STAT1 both at tyr 701 and ser 727 residues followed by STAT1 homodimer, which enter into the nucleus. Treatment of HTR-8/SVneo cells with IFNG also leads to an increase in the expression of BATF2 both at the transcript as well as protein levels. Panel A depicts role of STAT1 and BATF2 in IFNG-mediated decrease in invasion of HTR-8/SVneo cells by regulating expression of JUN and also regulate each-other's expression. Panel B represents the increase in invasion of HTR-8/SVneo cells subsequent to IFNG treatment after silencing of both STAT1 and BATF2, respectively. The increase in invasion, subsequent to silencing of STAT1 and BATF2 may be due to the upregulation in the expression of JUN, which in turn promotes AP-1 transcription factor by binding to its promoter site thereby increases invasion of HTR-8/SVneo cells.