Figures & data
Table 1. Subunits of the BRM chromatin-remodeling complex interact with CLK in the nucleus of Drosophila Schneider (S2) cells as detected by FLAG affinity purification followed by mass spectrometry.
Figure 1. The role of BRM in Drosophila circadian transcription. (A) Flies in which brm RNAi is used to knock down brm expression are expected to exhibit both decrease in catalytic and non-catalytic activity. Decrease in the latter is postulated to result in decreased recruitment of BRM-interacting proteins to CLK-regulated promoter regions. Interacting proteins are designated as Px, Py, and Pz. Reduction in catalytic activity results in decrease in nucleosome density. (B) Flies expressing the brmK804R allele express a catalytically-inactive BRM protein that is unable to hydrolyze ATP to remodel chromatin (increase nucleosome density), yet retains interactions with other proteins.Citation43 (C) Control “wild type” flies expressing native BRM protein with intact catalytic and non-catalytic activity. (D) An overview of findings from Kwok et al,Citation36 including the phenotypes of the two brm mutants with respect to circadian locomotor activity rhythm, expression of core clock genes, nucleosome density, and CLK localization at the per promoter.
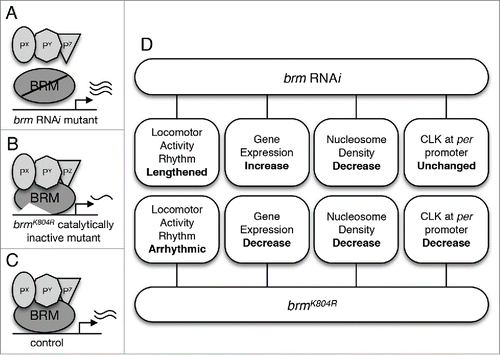
Figure 2. The effects of two classes of brm mutants on CLK-activated transcription. (A) Knockdown of brm via RNAi results in the reduction of both catalytic and non-catalytic functions. The decrease in catalytic activity leads to lower nucleosome density as compared to control, and decrease in non-catalytic function impairs recruitment of repressive complexes. As a result, CLK-activated gene expression is elevated. (B) Expression of catalytically-inactive BRMK804R protein results in decreased nucleosome density. The more open chromatin allows for an augmented effect of the non-catalytic function retained by BRMK804R, which may include recruitment of repressors. Experimental data suggests that interactors of BRM have a negative effect on CLK binding to the per promoter, and per expression is downregulated as a result of decreased CLK binding.Citation36 (C) Wild type BRM possesses both non-catalytic and catalytic functions that balance each other and serve to fine-tune CLK-activated transcription. Non-catalytic function of BRM may promote recruitment of proteins that have repressive effects on transcription. The over-recruitment of repressive factors is prevented by the catalytic function of BRM to maintain nucleosome density.
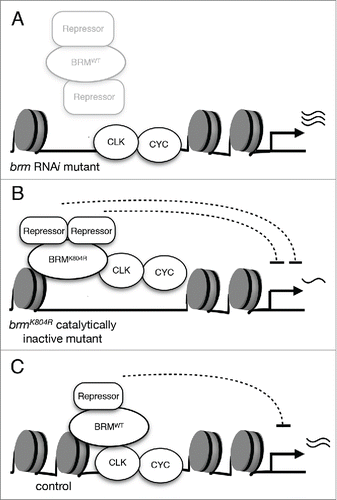
Figure 3. The interactions of BRM with additional factors regulate its recruitment to promoter regions and modulate circadian transcription. (A) The regulation of circadian gene expression in Drosophila is likely orchestrated through the interaction of a chromatin remodeler such as BRM with multiple epigenetic modifiers. Examples of candidate BRM-interacting repressors are depicted. (B) The possible role of CLK as well as PER and TIM in the recruitment and localization of BRM to the per promoter.
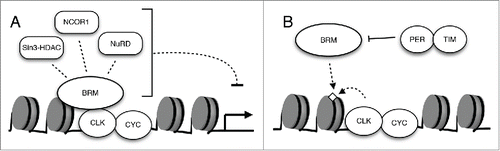
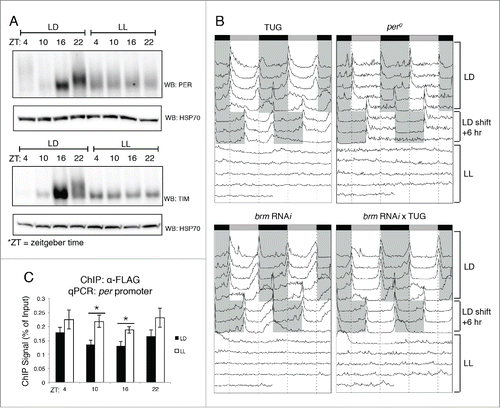
Figure 4. Disruption in PER and TIM expression using constant light treatment (LL) affects BRM localization at circadian promoters. (A) Verification that circadian rhythms in PER (top panel) and TIM (bottom panel) accumulation and post-translational modifications are abolished in LL conditions. Protein expression patterns in control flies that were entrained for 3 d in 12hr light: 12 hr dark conditions at 25°C are compared to flies subjected to constant light conditions (LL) for one day following 3 d of LD entrainment. Protein was extracted from fly heads as described in Kwok et al.Citation36 and resolved by SDS-PAGE. Antibodies against PER and TIM have been previously described.Citation36 Loading was normalized using anti-HSP70 (Sigma, St. Louis, MO). (B) Locomotor activity rhythms verify loss of rhythmicity in LL conditions. Male flies were subjected to locomotor activity assays using the Drosophila Activity Monitoring System (DAMS) (Trikinetics, Waltham, MA) as described in Chiu et al.Citation44 Flies were entrained for 4 d in 12hr light: 12 hr dark conditions. On the fifth day, LD conditions were shifted +6 h to confirm that flies did not exhibit entrainment defects. Starting the 8th day, flies held in LL conditions and periodicity of the clock was assessed. Shaded areas = lights-off. TUG = tim-(UAS)-Gal4 driver.Citation70 (C) Chromatin immunoprecipitation (ChIP) assay showing BRM localization to the per promoter in LD vs. LL conditions. Transgenic flies expressing BRM-FLAG driven by TUG were harvested at the 4 indicated time points (ZT) in either LD or LL conditions. α-FLAG (Sigma, St. Louis, MO) was used in ChIP-qPCR experiments as described in Kwok et al.Citation36 Data shown are from 3 biological replicates with technical triplicates performed during qPCR. Error bars = SEM for biological replicates (n = 3). Two-tailed t-tests were used to determine statistical differences (P < 0.05) between LD and LL treatments at each ZT. Asterisks denote significant difference observed (P < 0.05). Experimental procedures for ChIP-qPCR are detailed in Kwok et al.Citation36