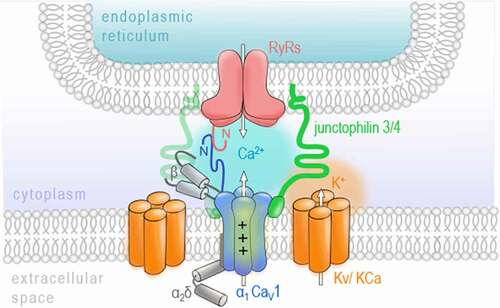
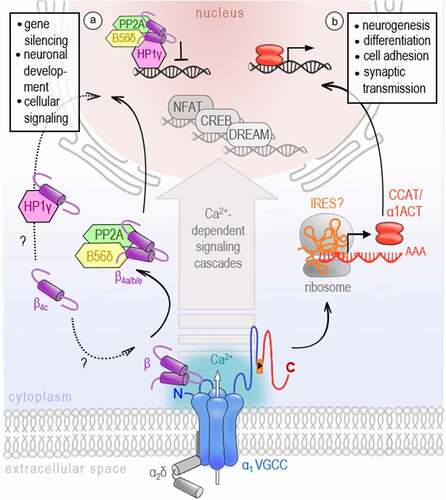
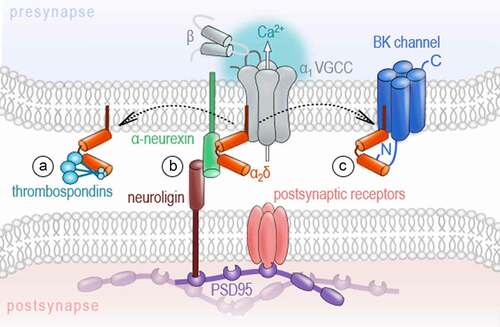
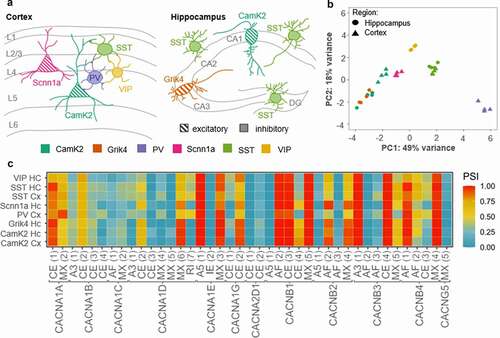
A) Schematic overview representing different neuronal cell populations included in the RNA sequencing data (data is publicly available; accession code: GSE133291
[Citation110]). The scheme was adapted from Furlanis and colleagues. The color-code for the cell types is used in B. CamK2= Ca
2+/calmodulin-dependent protein kinase II; Grik4= glutamate receptor, ionotropic, kainite 4; PV= parvalbumin; Scnn1a=α subunit of the epithelial sodium channel ENaC; SSTsomatostatin; VIP- vasoactive intestinal peptide. B) The principal component analysis (PCA) of the gene expression demonstrates a differential regulation of VGCC genes across excitatory and inhibitory neuronal cell types. Especially, the cluster of PV interneurons localizes with asignificant gap to other neuron types on principal component 1, which explains roughly fifty percent of the observed variance. Data analysis: The fastq-files were mapped to the mm10 reference genome using STAR
[Citation117]. The PCA was performed with DESeq2 in R
[Citation118,Citation119] and plotted with ggplot2
[Citation120]. C) Heatmap of alternatively spliced transcripts of VGCC genes. We have analyzed the occurrence of the following splice events: cassette exon (CE), mutually exclusive exon (MX), retained intron (RI), alternative 5’ splice-site (A5), alternative 3’ splice-site (A3) and alternative first exon (AF), schematically shown in the lower part of C. Only those events that show a significant variance between the samples (ANOVA p-value < 0.01) were included. Overall, a difference between the PV neurons and the remaining samples in the splice patterns for the VGCC genes can be observed (e.g.
CACNA1A MX(2),
CACNB1 CE(4) or
CANB3 MX (5)). Since the exon numbering depends on the transcript variant of a respective gene, the exact position for the splice variants (for instance, the start and end coordinates of the included exon) of the heatmap are listed here:
CACNA1A: CE (1) chr8:84601756−84601821, MX (2) chr8:84614695−84614791,
CACNA1B: A3 (1) chr2:4718122, CE (2) chr2:24642853−24642858, CE (3) chr2:24656711−24656722, CE (4) chr2:24682976−24683038,
CACNA1C: AF (1) chr6:119196231−119196093, MX (2) chr6:118637730−118637813,
CACNA1D: A3 (1) chr14:30129848, CE (2) chr14:30129789−30129848, CE (3) chr14:30137041−30137124, MX (4) chr14:30089296−30089379, MX (5) chr14:30107653−30107712, MX (6) chr14:30171296−30171399, RI (7) chr14:30128798−30129789,
CACNA1E: A5 (1)chr1:154471338, CE (2) chr1:154404885−154405013, MX (3) chr1:154416061−154416157,
CACNA1G: CE (1) chr11:94423671−94423724, CE (2) chr11:94439709−94439777,
CACNA2D1: CE (1) chr5:16322539−16322595, CE (2) chr5:16341990−16342010,
CACNB1:A5 (1) chr11:98010004, AF (2) chr11:98023034−98022887, CE (3) chr11:98010627−98010646, CE (4) chr11:98011343−98011497, MX (5) chr11:98010627−98010646,
CACNB2: A5 (1) chr2:4971646, AF (2) chr2:4739216−14739763,AF (3) chr2:4763129−14763992, MX (4) chr2:14967942−14968075,
CACNB3: A3 (1)chr15:98640686, AF (2) chr15:98631805−98631931, AF (3) chr15:98632376−98632520, CE (4) chr15:98640959−98640978, MX (5) chr15:98640959−98640978,
CACNB4: AF (1)chr2:52676582−52676271, AF (2) chr2:52676831−52676271, CE (3) chr2:52556202−52556361, MX (4) chr2:52465894−52465913,
CACNG5: AF (1) chr11:107915055−107914900. For this analysis, the fastq-files were mapped to the referencegenome (mm10) with Salmon
[Citation121] and the percentage-spliced in (PSI) values were computedusing SUPPA2
[Citation122].