Figures & data
Table 1. Comparison of semen parameters between men with normal (n=32) and abnormal (n=23) semen parameters.
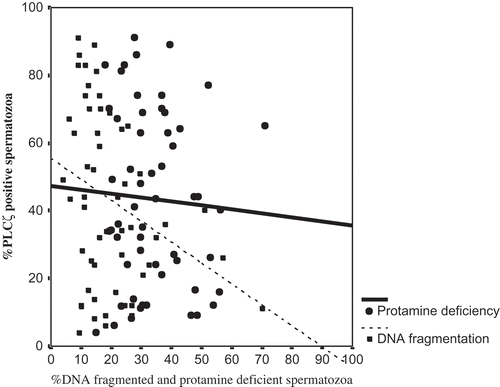
Figure 1. Comparison of mean percentage of sperm PLCζ, SDF, and protamine deficiency between men with normal and abnormal semen parameters. The mean percentage of PLCζ positive spermatozoa was significantly lower, and percentage of SDF and protamine deficient spermatozoa were significantly higher in men with abnormal semen parameters compared to men with normal semen parameters, respectively. SDF: sperm DNA fragmentation; PLCζ: phospholipase C-zeta. # shows significant difference between the two groups at p<0.05.
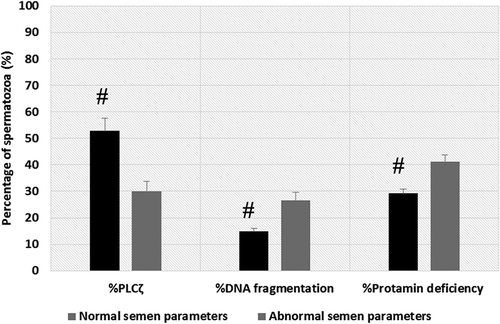
Figure 2. Flow cytometry analysis of percentages of PLCζ positive spermatozoa in two semen samples with normal (A) and abnormal (B) semen parameters. Percentage of PLCζ positive spermatozoa was higher in individuals with normal parameters compared to abnormal parameters. Fluorescence images of spermatozoa stained with TUNEL (C) and CMA3 staining (D) for DNA fragmentation and protamine deficiency, respectively. Spermatozoa with green staining (TUNEL+) and bright yellow staining (CMA3+) were considered as protamine deficient and DNA fragmented, respectively. PLCζ: phospholipase C-zeta; CMA3: chromomycin A3; TUNEL: terminal deoxynucleotidyl transferase dUTP nick end labelling.
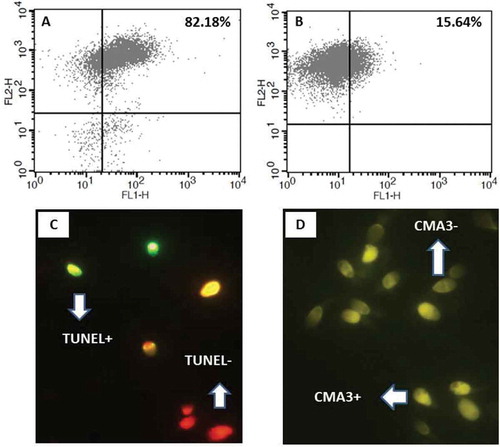
Table 2. Correlation between percentage of spermatozoa presenting DNA fragmentation, protamine deficiency, and PLCζ with sperm concentration, sperm motility, and sperm abnormal morphology (n=55).
Table 3. Multiple regression analysis between sperm parameters and percentage of PLCζ positive spermatozoa.