Figures & data
Figure 1. Causative variant detection in this family. (A) Pedigree of the MFS family reported in this study. Squares and circles indicate males and females, respectively. The black symbols II-1 (the proband) and II-2 (his younger brother) represent the male patients who suffered from MFS. The blank symbols show I-1 (his father) and II-3 (his wife) were unaffected and I-2 (his mother died of heart disease without specific diagnosis). (B) The pathogenic mutation site (FBN1: NM_000138; chr15:48,736,736 c.6037 + 2 T > C) of II-1 (the proband) detected by NGS. The figure provides the result of the antisense chain reads. (C) Sanger sequencing results of the FBN1 gene in the MFS family. II-1 and II-2 carried the FBN1 gene mutation c.6037 + 2 T > C, while I-1 and II-3 were wild-type (WT)
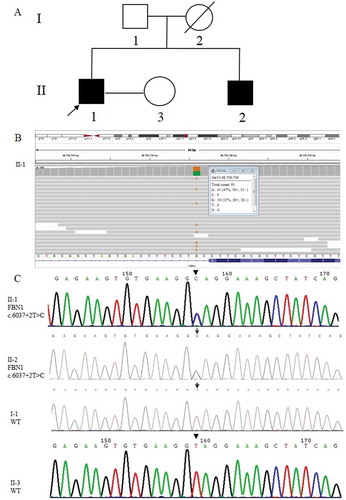
Table 1. Results of preimplantation genetic testing of 14 biopsied samples
Figure 2. Haplotyping results. 60 informative polymorphic SNP markers located upstream and downstream of the pathogenic site were selected to establish the haplotype. Ten of these SNPs (rs48523284, rs48540199, rs48564160, rs48584883 and rs48633092 are located upstream of the pathogenic site; rs48967277, rs49247626, rs49290738, rs49338132 and rs49359620) are selected for this figure. Diagram showing the results of haplotype analysis of the male, female and 14 blastocysts (E1-14). E3-8 and E13-14 inherited an affected haplotype (yellow-line allele) from the male, whereas E1, E2, and E9-12 inherited a normal haplotype (orange-line allele). ‘?’ indicates ADO or LDO
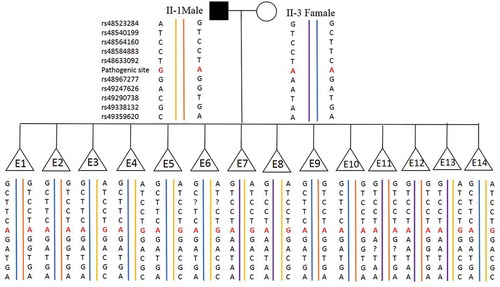
Figure 3. NGS-based SNP haplotyping results of mother (M), father (F), and 14 embryos (E1-E14). 60 informative polymorphic SNP markers located upstream and downstream of the mutation site in FBN1 were selected to establish the haplotype. The positions marked in green color are paternal informative SNPs while the positions in light green are maternal. The red squares in position 48,736,736 represent the mutation sites (G in red) and the yellow ones represent ADO sites. E1, E2, and E9-E12 exhibited an unaffected haplotype with the normal paternally inherited allele (F1), whereas E3-E7, E13, and E14 exhibited an affected haplotype pattern (F0). Question mark represents the undetected site which was actually a ‘Locus drop out’. Single solid line represents the gene boundary, while double solid line represents the pathogenic site boundary
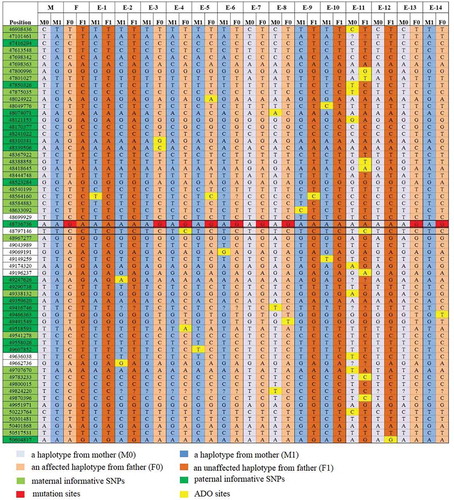
Figure 4. Copy number variation results of 6 samples (E1, E2, E9-12). CNV of the 6 embryos with normal results of Sanger sequencing and SNP haplotyping (E1, E2, E9-12) were shown both in and (sex chromosomes not shown), while the other 8 embryos with abnormal results of Sanger sequencing and SNP haplotyping (E3-8, E13, E14) were not tested for CNV. The maps shown that sample E1, E2, E9 and E10 were diploid, whereas E11 was mosaic [46, XX, del mosaic (2) (47.7%), del mosaic (4) (46.8%)] and E12 was aneuploid (47, XX, dup19)
![Figure 4. Copy number variation results of 6 samples (E1, E2, E9-12). CNV of the 6 embryos with normal results of Sanger sequencing and SNP haplotyping (E1, E2, E9-12) were shown both in Table 1 and Figure 3 (sex chromosomes not shown), while the other 8 embryos with abnormal results of Sanger sequencing and SNP haplotyping (E3-8, E13, E14) were not tested for CNV. The maps shown that sample E1, E2, E9 and E10 were diploid, whereas E11 was mosaic [46, XX, del mosaic (2) (47.7%), del mosaic (4) (46.8%)] and E12 was aneuploid (47, XX, dup19)](/cms/asset/caa4ee28-dd3b-45e9-87cd-bbc67b956203/iaan_a_1926574_f0004_oc.jpg)
