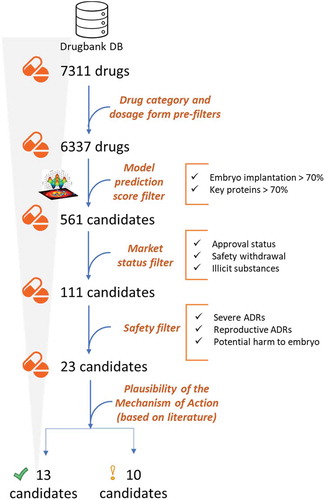
Figures & data
Table 1. EI molecular characterization
Table 2. List of the key proteins in successful EI identified by the mathematical models
Figure 1. Visual representation of the physiological motives involved in EI. Proteins reported to be associated with each of the motives (according to scientific bibliography) are indicated
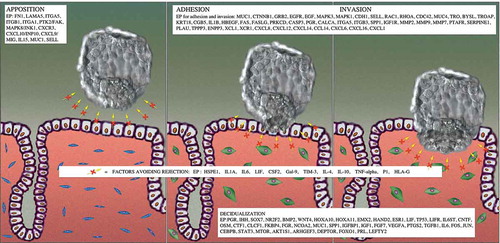
Figure 2. Steps followed to generate mathematical models of EI. First, proteins with a known role in EI (namely protein effectors) were determined by literature search. An exhaustive protein network was grown around these proteins incorporating information from several scientific databases. The protein network was then trained with functional information in humans and two types of mathematical models were generated, one mimicking successful EI and another simulating unsuccessful EI after IVF (this one trained with endometrial fluid-proteomics data from women collected immediately before embryo transfer in IVF cycles, linked to a known pregnancy outcome (pregnant or non‐pregnant). The analyses of these models through two different mathematical approaches (triggering analysis and model reversion analysis) allowed the identification of key proteins in successful EI
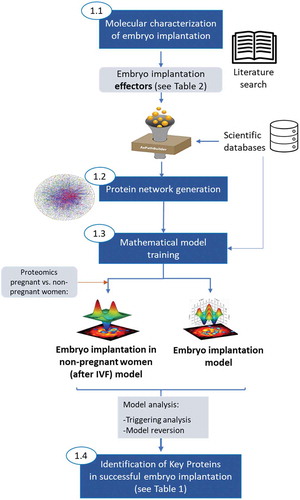
Table 3. List of repositioned candidates