Figures & data
Figure 1. Schematic diagram of an IgG-Fab BsAb (A). Diagrams to the right of the correctly assembled IgG-Fab are potential mispairings related to the lack of LC specificity for a particularly HC Fd region. (B) Schematic diagram of the domain architecture of a α/β TCR. The receptor is a heterodimer consisting of 2 chains that each comprise a V-class and a C-class Ig-fold, much like an immunoglobulin Fab. (C) Superposition of the structures of an IgG1 CH1/Cκ heterodimer (pdb id: 3HC0) and the constant domains of an α/β TCR (pdb id: 3ARB). The structural homology between Cβ and Cκ as well as that between Cα and CH1 is apparent. (D) Diagram demonstrating the exchange of the CH1/Cκ domains with the TCR Cα/Cβ domains within an IgG1 antibody (denoted IgG_TCR).
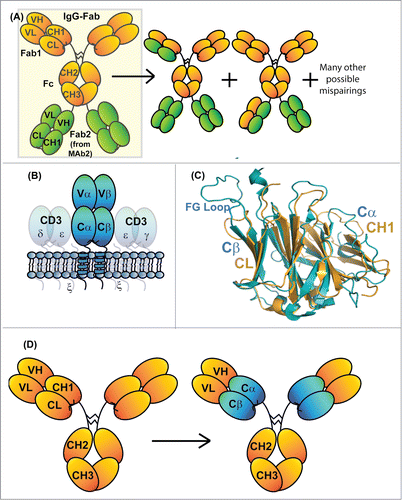
Table 1. Biochemical characterization of initial IgG_TCRs
Figure 2. Characterization of initial IgG_TCR constructs. (A, B) SDS-PAGE analysis of IgG_TCR constructs under reducing, 10 mM DTT, (A) and non-reducing conditions (B). 15 μL of protein G magnetic bead purified protein from 2 mL 293F purifications was added to each lane. β1 and β2 differ in the N-terminal residues of the β-domain (β1 starts with E117, while β2 starts with K121). Similarly, α1 and α2 differ in the N-terminal residues of the α-domain (α1 starts with P116, while α2 starts with I118). (C) Non-reduced (left side of gel) and reduced (right side of gel) SDS-PAGE analysis of protein G pull-downs from supernatants expressed using mismatched and matched pairs of IgG and IgG_TCR heavy and light chains. (D) Cation exchange separation of IgG_TCR proteins secreted with 0 (1st peak), 1 (middle peak), or 2 (3rd peak) associated LCs. The inset shows the SDS-PAGE analysis of the 3 cation exchange fractions. (E) Binding activity of the protein fractions separated in (D), demonstrating the importance of LC association for binding to antigen. The association of the IgG_TCR protein can be observed between 300–600 s, while the antigen (IL-17 in this case) association can be observed between 800–1000 s. (F) Improvement in the uniform expression of fully paired (HC2LC2) IgG_TCR proteins after truncating the C-terminal tail of the β-constant domain of the LC. HC2LC2 elution time was at 13.5 minutes based on static light scattering analysis.
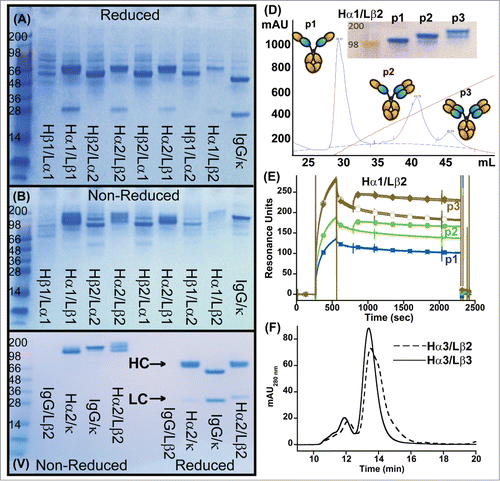
Table 2. Occupancy of predicted N-linked glycosylation sites
Figure 3. Effect of deleting N-linked glycosylation on HEK293 expression of IgG_TCR proteins. Reduced SDS-PAGE analysis (A) and analytical SEC (B) of fully glycosylated (WT) IgG_TCR, single N-linked glycosylation deletion mutants, double mutants, and a triple mutant after protein G pull-down from 2 mL HEK293 expression supernatants. (C) DSC analysis of fully glycosylated (WT) and single N-linked glycosylation deletion mutants of IgG_TCR proteins after 100 mL HEK293 scale-up and protein A purification.
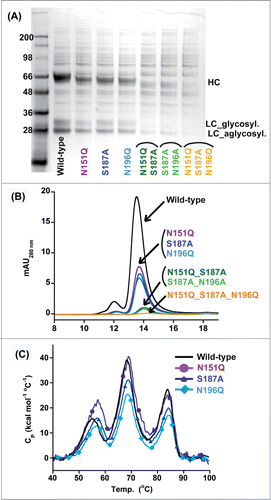
Figure 4. Replacement of the FG loop from the β-constant domain with common β-turn motifs. (A) Stick diagram of the structure of the β-constant domain (from pdb 3QEU) where the FG loop is colored orange. Non-reduced (top) and reduced (bottom) SDS-PAGE analyses (B) and analytical SEC (C) of WT IgG_TCR, FG loop-deleted IgG_TCR, and IgG_TCRs with the FG loop replaced with a PS (proline_serine, Type I), NG (Type I’), and GN (Type II’) β-turn. The analyses in (B) and (C) were performed on IgG_TCR proteins expressed at the 2 mL scale in HEK293 and pulled down using protein G magnetic beads. (D) DSC analyses of WT IgG_TCR and FG loop replaced IgG_TCR after scale-up and protein A purification.
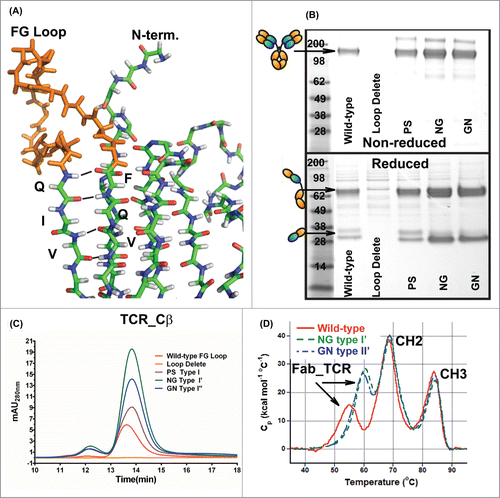
Figure 5. Biophysical characterization of HER2×HER2 IgG-Fab BsAbs produced using IgG_TCR modalities to direct LC assembly. Panels A and B are non-reduced (left) and reduced (right) SDS-PAGE analysis and analytical SEC, respectively, of C-BsAb and N-BsAbs. Panels E and F are an evaluation of the HER-2 binding properties of trastuzumab IgG_TCR, pertuzumab IgG_TCR, C-BsAb, and N-BsAb by analyzing their ability to block 40 nM HER-2 from binding surfaces labeled with IgG1 trastuzumab (C) or pertuzumab (D). Panels E and F are intact mass spectrometry analyses of C-BsAb and N-BsAb, respectively under reducing conditions. The N-BsAb contained the VL_Y36F mutation and VL_Q38D/VH_Q39K to reduce the affinity of the trastuzumab LC for the pertuzumab Fd containing Cα/Cβ. The spectra show the levels of LC within the IgG-Fab BsAbs. The HC was heavily N- and O-glycosylated; therefore, non-reduced spectra were complex.
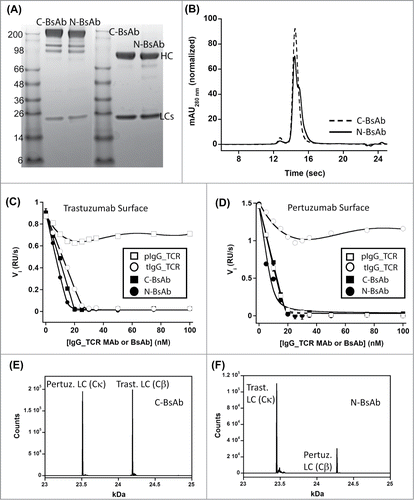
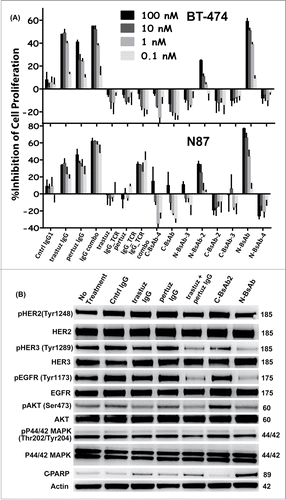
Figure 6. Biological activity of IgG_TCR BsAbs. The effect of IgG_TCR BsAbs on the FBS-driven proliferation of (A) BT-474 breast cancer cells (top) and N87 gastric cancer cells (bottom). (B) Western blot analyses of the phosphorylated state of EGFR, HER-2, HER-3, Akt, and Erk from N87 tumor cells grown for 48 hours in FBS in the presence of various anti-HER-2 monoclonal and bispecific antibodies. Additionally, the presence of cleaved PARP was evaluated on the blot. Actin was probed to demonstrate the normalized amount of protein loaded into each well.