Figures & data
Figure 1. Affinity determination of the rFcRn/rIgG2a interaction at pH 5.8 using various assay orientations. Kinetic analysis of rIgG2a flowed over biotinylated rFcRn that was captured at (A) high or (B) low capacities via amine-coupled neutravidin on a Biacore C1 chip; kinetic analysis of rFcRn flowed over (C) antigen-captured rIgG2a on a ProteOn NLC chip or (D) amine-coupled rIgG2a on a Biacore CM5 chip; and (E) solution affinity using a high capacity of amine-coupled rIgG2a on a Biacore CM5 chip to probe for free rFcRn in equilibrated mixtures with titrating levels of rIgG2a. Analytes were injected as a 3-fold dilution series with top at 500 nM (A and B) or as both a 3-fold (green) and 5-fold (red) dilution series with top at 1000 nM (C) or 3000 nM (D); the 3000 nM curves have been excluded from D. Panel E shows the titration of 17 nM rFcRn with 2 unrelated mAbs of subtype rIgG2a (distinguished by the solid or open symbols). All samples were analyzed in replicate binding cycles. Each panel shows an example data set (N of 1) where the measured data (colored lines) were fit globally to a simple model (black lines). The KD values are the mean ± SD for N independent measurements. See Table 1.
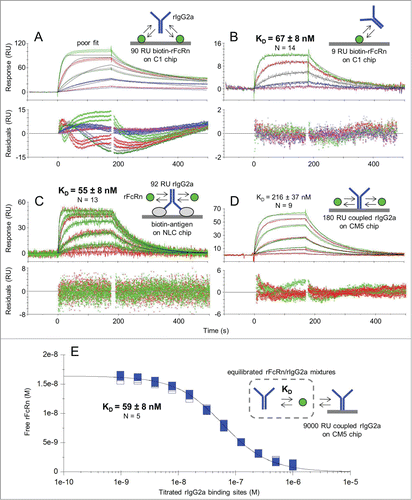
Table 1. Affinity determination of the rFcRn/rIgG2a interaction at pH 5.8 using 3 different assay formats on SPR platforms. Parameter values represent the mean ± SD of N independent measurements on 5 unrelated mAbs. The range of KD values obtained is also shown in parenthesis. ND = not determined
Figure 2. Kinetic analysis at pH 5.8 of a multi-species panel of FcRn proteins binding as analytes to trastuzumab hIgG1-N434Y that was first captured via (A) biotinylated mFcRn or (B) biotinylated hErbB2 on Biacore C1 chips to which neutravidin was amine-coupled. Analytes were injected in duplicate binding cycles as a 3-fold dilution series with top at 180, 300, 60, or 100 nM for hFcRn, cyFcRn, mFcRn, and rFcRn respectively. Each overlay plot shows a representative example of the measured data (colored lines) and the global fit to a simple model (black lines) for a typical experiment (N of 1) and the KD values are the mean ± SD of N = 6.
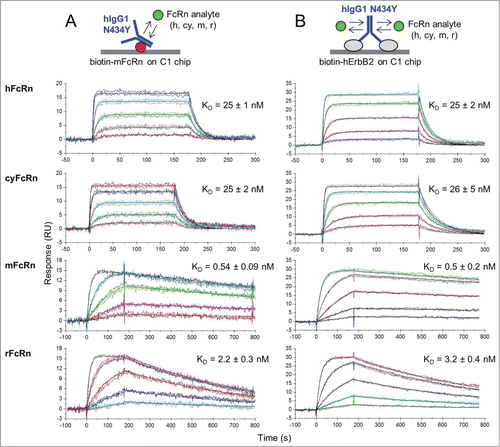
Figure 3. One-shot kinetic analysis at pH 5.8 of mFcRn binding as analyte to (A) trastuzumab homodimers and (B) their corresponding heterodimers that were captured at similar levels via ProteOn NLC or GLC chips coated with biotinylated hErbB2. Mouse FcRn was injected as a 3-fold or 5-fold dilution series with top concentrations of 180 nM (WT and N434H) or 85 nM (N434Y). Each overlay plot shows a representative example of the measured data (noisy lines) and the global fit (smooth lines) for a subset of the constructs tested. Multiple constructs per variants were fit simultaneously to give global apparent mean KD ± SD (N = 4) values of 58 ± 10 nM (WT-hIgG1, WT:WT, WT E arm, WT R arm, I253A:WT and AAA:WT), 5 ± 1 nM (N434H, N434H R arm, I253A:N434H and AAA:N434H), and 0.47 ± 0.07 nM (N434Y, N434Y R arm, AAA:N434Y). For a summary of all interactions tested in this way, see Supplemental Material (Table S2).
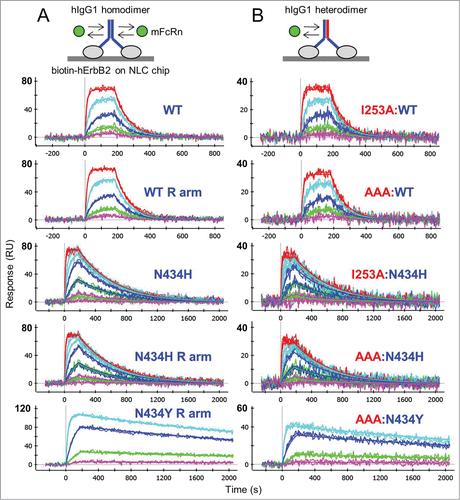
Table 2. Summary of the apparent affinities determined in 3 different assay formats for the interactions of mFcRn and rFcRn with trastuzumab variants. Reported KD values (nM) are the mean (± SD) of N independent measurements. The IgG kinetic data are a subset of those shown in Table S1, where hIgG1 heterodimers were flowed as monovalent analytes over biotinylated FcRn on ProteOn GLM sensor chips. The FcRn kinetic data are a subset of those shown in Table S2, where FcRn was flowed as analyte over hErbB2-captured trastuzumab on ProteOn NLC or GLC sensor chips
Figure 4. Solution affinity determinations of mFcRn binding to trastuzumab hIgG1 variants. (A) WT and AAA:WT at pH 5.8, (B) N434H and AAA:N434H at pH 5.8, (C) N434Y and AAA:N434Y at pH 5.8 and (D) N434Y and AAA:N434Y at pH 7.4. Each overlay plot shows an example of the KD-controlled curves obtained for titrations of a hIgG1 homodimer (blue symbols) and its corresponding heterodimer (red symbols) into a fixed concentration of mFcRn (4.5, 2.7, 0.9, and 36 nM for panels A-D respectively). See .
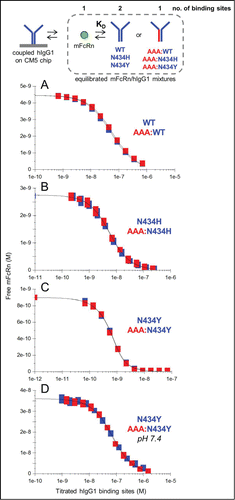
Table 3. Affinities for a multi-species panel of FcRn/IgG interactions determined at pH 5.8 at both 25°C and 37°C. These data were generated in a series of one-shot kinetic experiments by flowing FcRn over antigen-captured, anti-Id Fab-captured, or biotinylated IgGs on ProteOn NLC chips. The KD values (nM) represent the mean (± SD) of N independent measurements. The hIgG1 category includes hIgG1 Fc stump (no variable region) and the hIgG2 category includes hIgG2Δa. NB = no binding (or barely binds) at 6 μM. NT = not tested
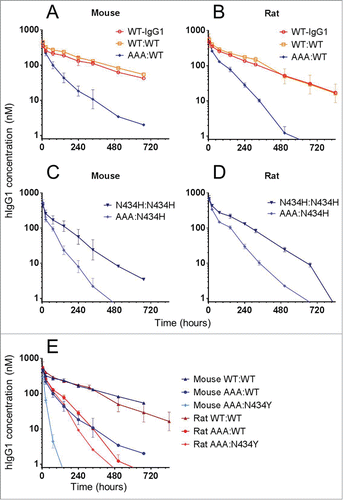
Figure 5. In vivo clearance of a panel of trastuzumab hIgG1 homodimers and heterodimers in mice and rats. Overlay plot of WT-hIgG1, WT:WT and AAA:WT in (A) mice and (B) rats. Overlay plot of N434H:N434H and AAA:N434H in (C) mice and (D) rats. (E) Overlay plot of WT:WT, AAA:WT, and AAA:N434Y in mice and rats. Data points represent the mean ± SD of 3 or 4 animals per group.