Figures & data
Figure 1. Deconvoluted mass spectra of the reduced antibody heavy chain(A and B) and Fc fragment generated by IdeS (C and D).
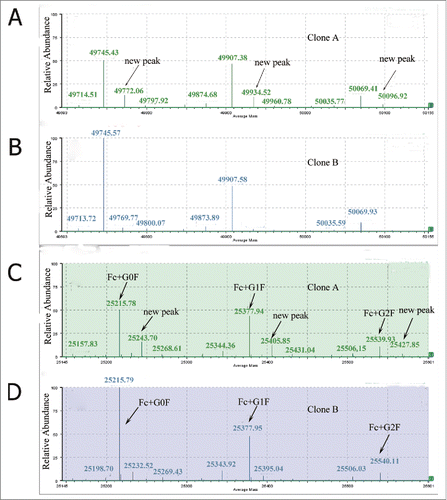
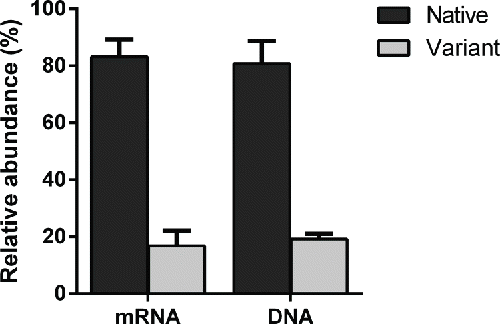
Figure 2. The tryptic peptide maps of Clone A and Clone B. The new peak (see arrow) at 49.10 min in Clone A is absent from Clone B.
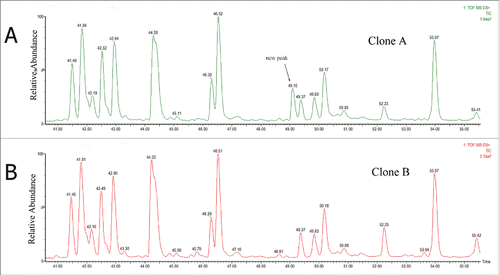
Figure 3. MS/MS spectra of precursor ions at (A) m/z of 701.56 for native peptide WQEGNVFSCSVMHEALHNHYTQK; and (B) m/z of 708.81 for variant peptide; and (C) m/z of 708.81 for synthetic peptide WQEGNVFSCSVMHEVLHNHYTQK.
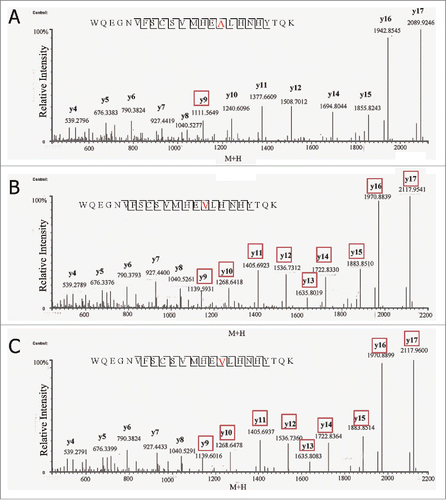
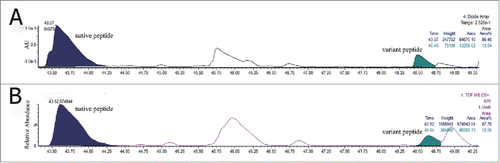
Figure 4. Total ion chromatogram of native peptide, WQEGNVFSCSVMHEALHNHYTQK and variant peptide, WQEGNVFSCSVMHEVLHNHYTQK in (A) non-spiked antibody tryptic digest and (B) spiked in WQEGNVFSCSVMHEVLHNHYTQK synthetic peptide.
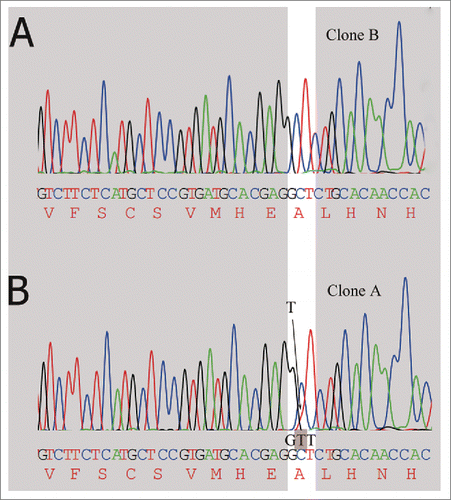
Figure 5. Detection of alanine codon (GCT) change to valine codon (GTT) in mRNA encoding the high chain by DNA sequencing. (A) The clone A has no mutation at the highlighted position, only the expected nucleotide C. (B) Small amount of mutated T (shaded in the trace) is detected along with the wild type nucleotide C.