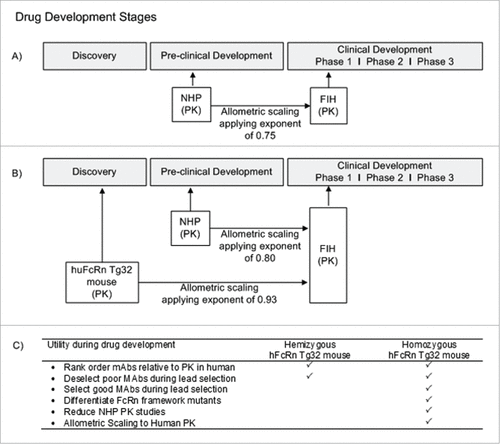
Figures & data
Figure 1. Panel of mAbs Used in Study. Interspecies CL for therapeutic mAbs. Panels A, B and C depict the 27 mAbs examined in this study, including 21 Pfizer mAbs (•![]()
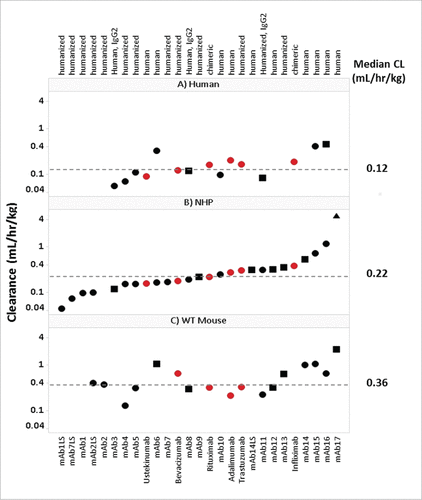
Figure 2. hFcRn Tg Mouse Strain Comparisons. PK profile results of Tg32 hemizygous and homozygous, Tg276 homozygous, WT C57Bl/6 and FcRn KO mice following administration of (A) mAb 14, (B) mAb 04 and (C) mAb17. (D) PK profile overlay in FcRn KO mice for mAb04 and mAb17. (E) PK parameters for mAb14, mAb04 and mAb17 in NHP and each mouse model. Data is depicted as the mean ± standard deviation for 3-6 animals/group for panels A-D.
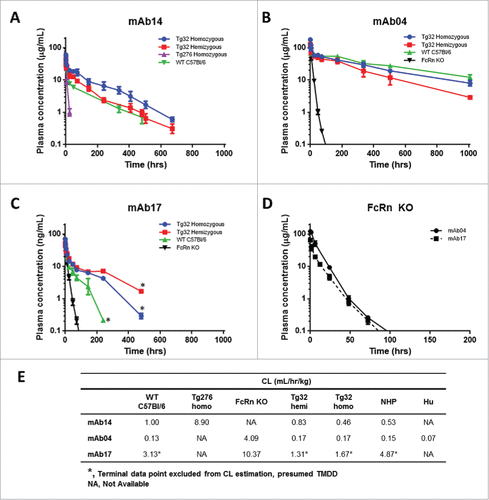
Figure 3. hFcRn Tissue Expression Profile in Tg32 Hemizygous and Homozygous Mice. Significant expression differences between homozygous and hemizygous genotypes were analyzed using an unpaired Mann-Whitney test where significance is indicated as single asterisk (*) for p < 0.1 and double asterisk (**) for p < 0.05.
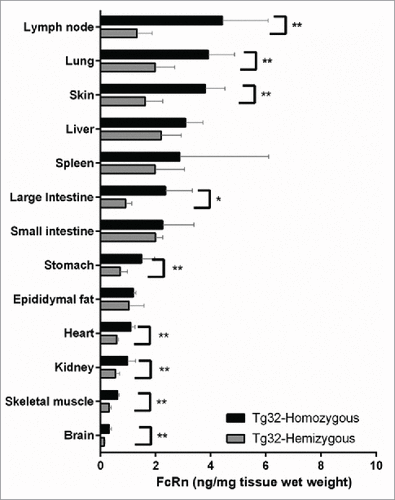
Figure 4. hFcRn Tg32 PK Differentiation in Framework Mutations. CL correlation between (A) Tg32 hemizygous and (B) homozygous mouse to NHP for 4 pairs of parent and FcRn+ mAbs (mAb01, mAb02, mAb07, mAb14) and (C) Summary of CL and T1/2 PK parameters for each parent and FcRn+ mAb in Tg32 hemizygous and homozygous mice and NHP. Statistical significance determined using a Student's paired t-test (*p < 0.05 and ** p < 0.01).
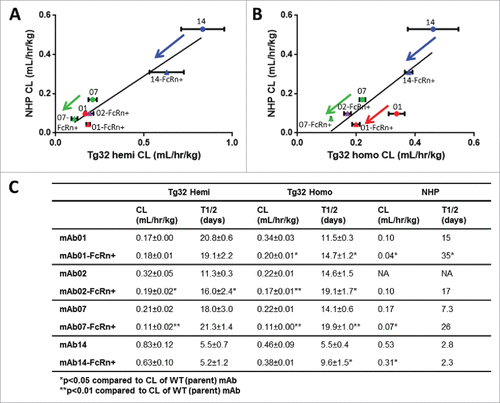
Figure 5. Correlation of mAb CL in Rodents to mAb CL in NHP. Linear correlation graphs of (A) WT mouse CL to NHP CL for 15/27 mAbs, (B) Tg32 hemizygous mouse CL to NHP CL for 25/27 mAbs and (C) Tg32 homozygous CL to NHP CL for 23/27 mAbs. Tg32 mouse CL results are shown as the mean ± standard deviation for 3-6 animals/group. Symbols: •![]()
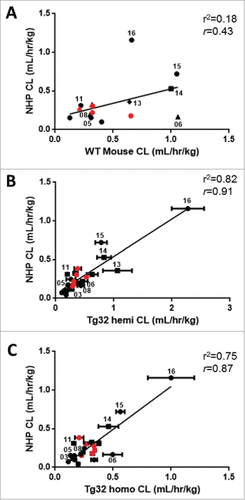
Figure 6. Correlation of mAb CL in Rodents vs Human and NHP vs Human. Linear correlation graphs of (A) WT mouse CL to human CL for 11/27 mAbs, (B) NHP CL to human CL for 15/27 mAbs, (C) Tg32 hemizygous CL to human CL for 15/27 mAbs and D) Tg32 homozygous CL to human CL for 15/27 mAbs. MAbs shown in panels B, C, and D represent the same 15 mAbs. Tg32 mouse CL results are shown as the mean ± standard deviation for 3-6 animals/group. Symbols: •![]()
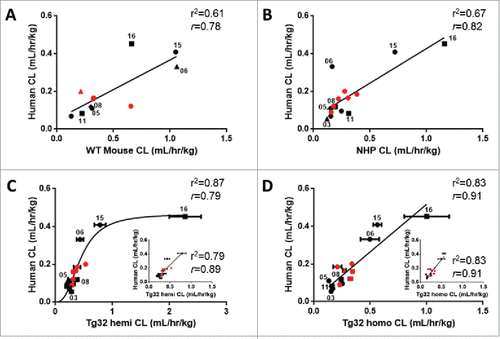
Figure 7. Single Species Allometric Scaling of CL for mAbs. Allometric Scaling for mAbs in NHP (15/27mAbs), WT mice (11/27mAbs), and hFcRn Tg32 hemizygous and homozygous mice (15/27 mAbs), comparing a standard scaling exponent of 0.75 (top row) to an empirically derived best fit exponent (middle row) per animal model. Results are plotted against a line of unity (solid black line) ± 2-fold error (dotted gray lines) where accuracy is described as the percentage (%) of mAbs predicted within 2-fold of the line of unity. Symbols: •![]()
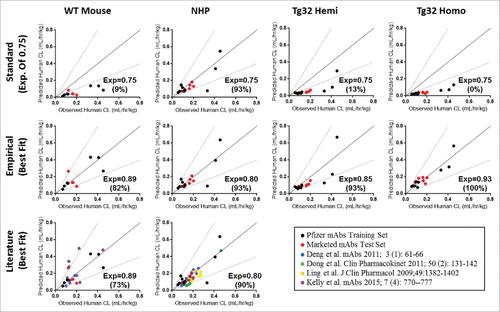
Figure 8. Utility of hFcRn Tg32 Mouse Model. (A) Current timeline for utility of NHP PK studies in preclinical development. (B) Suggested timeline for utility of hFcRn Tg32 PK studies, showing earlier use in discovery and application of allometric scaling using the exponent 0.93 to scale to human PK. (C) Use and selection of hemizygous vs homozygous Tg32 model.