Figures & data
Table 1. A comparison of the mass-confirmed glycans present at normalized abundances greater than 0.2% for innovator and biosimilar infliximab. In this report, we use the following glycan nomenclature: F- Fucose; G- Galactose; Sg- N-glycolylneuraminic acid, Ga- α-linked Galactose; A1- Monoantennary, A2- Biantennary. Numbers with parentheses indicate the preceding monosaccharide's linkage while those not in parentheses indicate the preceding characteristic's number. For example, F(6)A3G2Ga1Sg1 represents a core fucosylated triantennary glycan with 2 galactoses directly attached to antennae, 1 galactose linked via an α linkage, and one antenna terminated with an N-glycolylneuraminic acid. Symbols: blue square- N-acetylglucosamine; green circle- mannose, yellow circle- galactose, red triangle- fucose; gray diamond- N-glycolylneuraminic acid.
Figure 1. FLR chromatograms of released N-linked glycans from 3 innovators batches of infliximab (blue, black, and purple traces) and one batch of biosimilar (red trace). The symbols are the same as those used in .
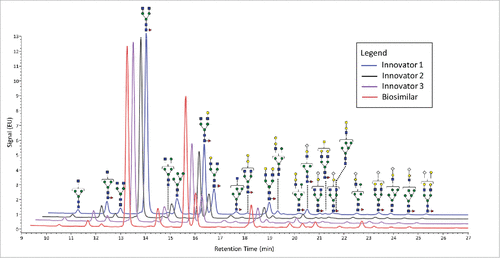
Figure 2. Differences in the normalized relative intensities (based on FLR data) for selected glycans featuring immunogenic glycans.
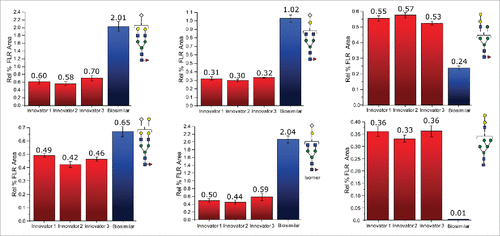
Figure 3. HDX MS deuterium exchange mass spectrometry comparability profile of the biosimilar and one of the innovator samples. (A) A butterfly plot of the average relative fractional exchange data for Innovator (top) versus biosimilar (bottom), as a function of peptide order. The x-axis is the calculated peptide midpoint. The y-axis is the average calculated relative fractional exchange. The orange, red, cyan, blue, and black lines correspond to data acquired at 30 s, and 1, 10, 60, and 240 min of deuteration, respectively, for both samples. The standard deviation between 2 measurements across all peptides was less than ±0 .05 Da, which is shown in the middle in gray. (B) Differential Plot.
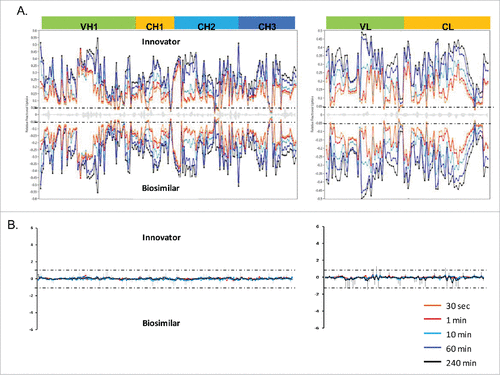
Figure 4. Comparison of deuterium incorporation of innovator and biosimilar samples. (A) Representative deuterium incorporation profiles of regions (residues 244–255, 245–254, 245–255, 285–303) in CH2 domains shows a minute difference. The red line represents the data from the biosimilar product, the green, cyan and blue lines represent the data from the 3 batches of the innovator samples. The experiments have been repeated in triplicate runs. (B) The location of the region that displayed minor difference among biosimilar and innovator samples are colored in red in the model structure of IgG1 (PDB: 1HZH). Glycosylation is shown in blue. Met255 is circled and shown in stick notation.
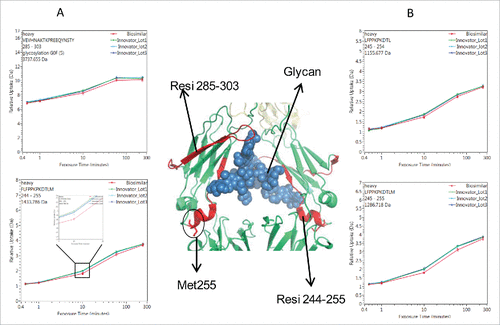
Figure 5. Representative peptides covered the complementarity determining regions (CDRs) of infliximab displayed identical conformation and dynamics. The heavy chain and light chain structures are colored in the 3D model of IgG1 (PDB: 1HZH) in green and yellow, respectively. The three light and heavy chain CDRs are colored in red. The deuterium incorporation curves of the sample peptides, which covered all the CDRs, are showed.
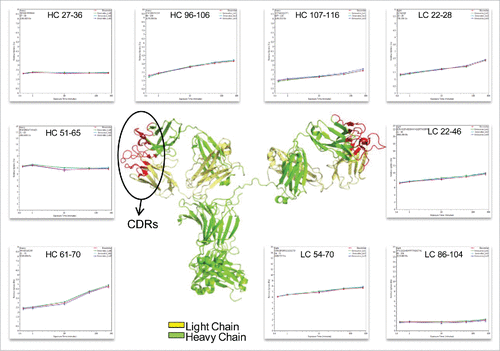
Table 2. A comparison of host cell proteins between innovator (A) and biosimilar (B) infliximab.