Figures & data
Figure 1. Pre-existing human antibodies to the Fab of human IgG1, IgG2 and IgG4. (A) X-ray crystal structure of the Fab region (PDB: 1HZH) including the upper hinge; light chain (gray), heavy chain (blue), interchain disulfide (orange), and upper hinge (magenta). In the isolated Fab molecule the upper hinge is a protruding unstructured region without structural and functional role. The residues of the upper hinge are displayed in magenta to indicate the T225L mutation (black) that is perturbing binding to pre-existing AHA. Numbering of residues is according to EU numbering nomenclature. (B) Pooled human serum was incubated with human IgG1 Fab with different upper hinge lengths and ends. Binding pre-existing antibodies detected by anti-Fc ELISA. Truncating the Fab C-terminus to D221 (D) and the C-terminal variant T225L (DKTHL) greatly reduced binding of pre-existing antibodies to almost background. Strong response is observed toward T223 as the C-terminal residue (DKT), coinciding with the cleavage site of human neutrophil elastase. The mean value of the individual data points is represented by the horizontal line. (C) Three different Fabs were incubated with pooled human serum and binding of pre-existing antibodies detected by ELISA. Significant signal is observed for different Fabs with DKTHT C-terminus. Reduced binding of pre-existing antibodies to the D221 and T225L C-terminus is detected across different Fabs. Fab-1 includes the antibody variable domain used in (B) and all other AHA binding experiments throughout this study. Fab-2 and Fab-3 are antibodies toward different antigens. (D) Pooled human serum was incubated with human IgG2 Fab and IgG4 Fabs with different upper hinge lengths and binding antibodies detected by ELISA. No pre-existing antibodies can be detected to the upper hinge of human IgG2 and IgG4.
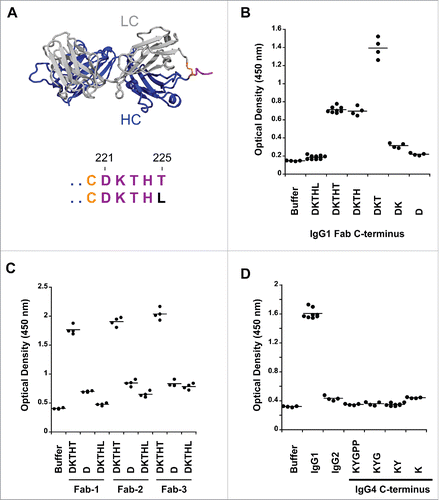
Figure 2. IgG1-2 chimera is inefficiently cleaved by IdeS. (A) Model of the F(ab′)2 region of antibody cAC10 modeled with MOE; light chain (gray), heavy chain (blue), interchain disulfide (orange), and lower hinge (magenta). The P1 position of IdeS is G236. Numbering of residues is according to EU numbering nomenclature. (B) Alignment of the lower hinge of IgG1 and the IgG1-2 chimera. Residues in cyan are IgG2 isotype residues introduced into the lower hinge of IgG2. (C) Cleavage efficiency of human IgG1 and IgG1-2 chimera. One mg/ml of IgG1 and IgG1-2 were incubated for 24 hours at 37°C with different IdeS amounts as indicated. Cleavage was analyzed by capillary electrophoresis. While IgG1 is efficiently cleaved into F(ab′)2 at an IdeS:IgG ratio of 1:500, IgG1-2 requires 50-fold higher IdeS concentrations for complete cleavage.
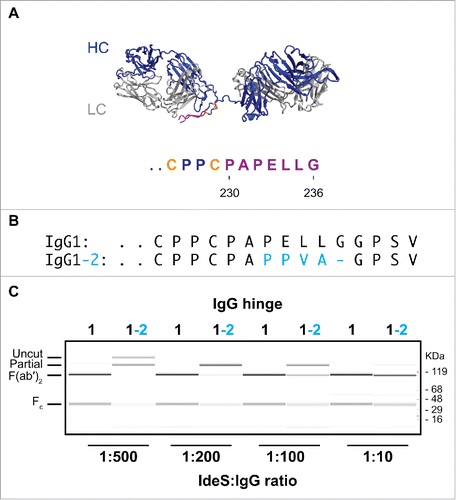
Figure 3. Cleavage of human IgG1 with variants in the P1 and P2 positions of IdeS. (A) Capillary electrophoresis of antibodies with variants at the P1 and P2 position were digested for 24 hours at 37°C with a 1:10 ratio of IdeS:IgG at 1mg/ml. The P1 and P2 residues are designated in 1-letter code. Leucine and glycine (L235G236) are the natural amino acids at these positions. All antibody variants can be completely cleaved into F(ab′)2 fragments. (B) The cleavage efficiency of the variants is assessed by the amount of F(ab′)2 produced at different IdeS:IgG ratios. While the variant VG is cleaved comparably to the wild-type sequence (LG), other variants require increased amounts of IdeS for complete digestion.
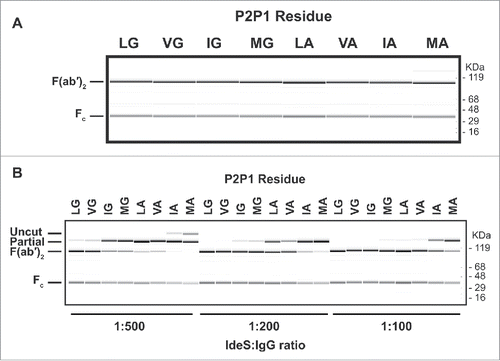
Figure 4. P1 and P2 variants do not remove recognition by pre-existing AHA. (A) IdeS is efficiently removed during purification and cannot be detected in the purified F(ab′)2 variants by SDS-PAGE followed by Coomassie staining (upper panel) or immunoblot analysis with anti-IdeS antibodies (lower panel). (B) Pooled human serum was incubated with human IgG1 Fab with T225 C-terminus and F(ab′)2 generated by IdeS cleavage of antibodies with variants in P1 and P2 positions. The pre-existing AHA binding was detected by ELISA. The hinge variants reduce reactivity to levels comparable to Fab, but do not eliminate reactivity completely.
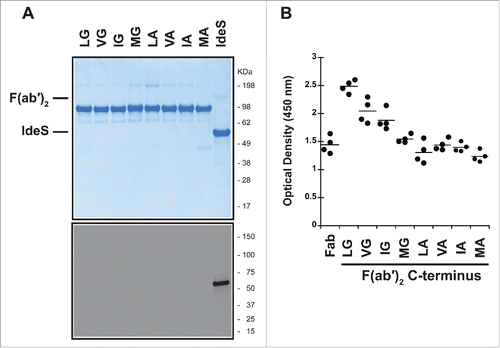
Figure 5. Truncating the lower hinge eliminates pre-existing AHA binding. (A) IdeS cleavage of antibodies with deletions in the IdeS P3, P4, and P5 sites. While deletion of the IdeS P3 residue (L234) in the lower hinge severely affects cleavage efficiency, deletion of the P4 (E233) or P5 (P232) positions do not affect cleavage with IdeS compared to wild-type (WT). (B) Deletion of the P4 and P5 positions is not sufficient to avoid binding of pre-existing AHA. (C) IdeS cleavage of antibodies with deletions of the IdeS P4 through P6 (ΔP456) and P4 through P7 (ΔP4567) sites. While cleavage efficiency of the ΔP4567 variant is slightly reduced compared to the wild-type lower hinge sequence (WT), ΔP456 displays cleavage efficiency comparable to the wild-type at IdeS:IgG ratio of 1:200. (D) Pooled human serum was incubated with F(ab′)2 produced by IdeS digestion and binding antibodies detected by ELISA. F(ab′)2 lower hinge deletions ΔP456 and ΔP4567 are not recognized by pre-existing AHA. (E) IdeS cleaves the ΔP456 hinge variant with high specificity. After digestion of wild-type (WT) and ΔP456 hinge IgG, reduced F(ab′)2 was analyzed by mass spectrometry. Only a single heavy chain species corresponding to the expected molecular mass was observed.
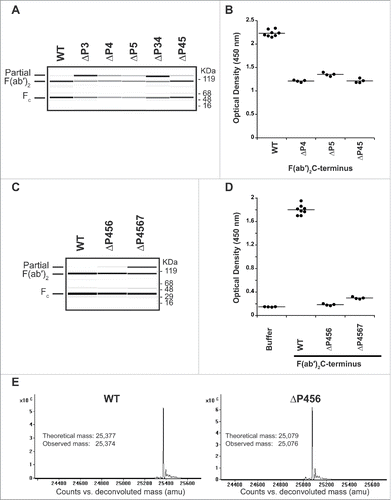
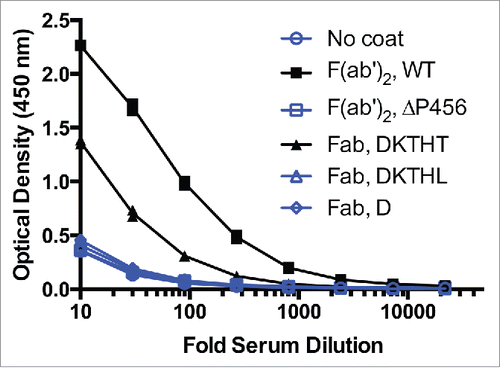
Figure 6. F(ab′)2 has stronger AHA binding than IgG1 Fab T225 in the AHA ELISA. Titration curves of F(ab′)2 and Fab variants in the AHA ELISA. F(ab′)2has 5-fold higher AHA reactivity than Fab T225. The dilutions corresponding to the OD (1.15) at the middle of the F(ab′)2 titration curves were 70 and 14 for F(ab′)2 and Fab, respectively. F(ab′)2, F(ab′)2 ΔP456, Fab T225, Fab T225L and Fab D221 were coated on the wells. Serial dilutions of pooled human serum were added to the wells and control wells were uncoated. Similar results were obtained in 4 other experiments. The data shown here and in and were collected from the same experiment.