Figures & data
Table 1. Molecule Properties and Impurity Level in post viral inactivation (VI) Protein A eluate neutralized (PAEN) pools.
Table 2. Adsorptive Depth Filters (ADFs) and adsorptive hybrid filters (AHFs) in lenticular format used for the study.
Figure 1. BSA Binding Capacity Performance of Adsorptive Filters. BSA binding capacity for adsorptive filters is shown over range of (A) conductivities and (B) flow rates. Error bars represent standard deviation of the mean (n = 3).
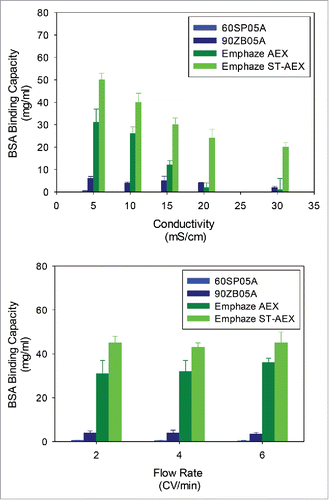
Figure 2. Post Viral Inactivation (VI) Neutralization pH Optimization. Neutralization to optimal pH after low pH hold for viral inactivation leads to selective precipitation of HCPs. Shown above are the recovery of mAb and HCP/DNA levels in viral inactivated and neutralized bulk samples post 0.2 µm filtration for mAb A.
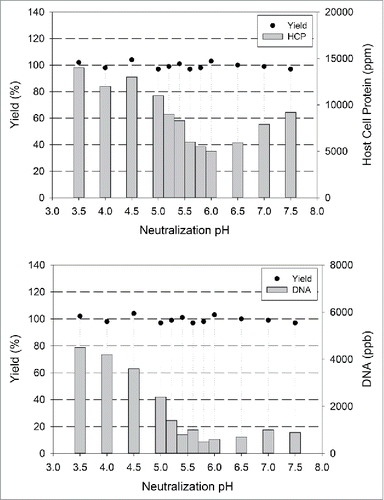
Figure 3. Resistance profiles as a function of throughput for neutralized mAb A bulk samples. Shown above are the resistance levels for neutralized bulk samples with different filters with 0.2 µm filtration as the refrence used for comparison. The data was collected at a filtration flux of 300 LMH for all the filters.
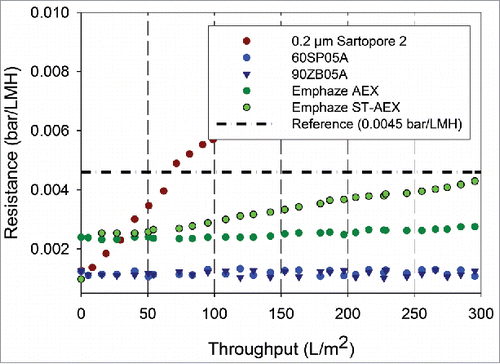
Figure 4. Breakthrough profiles for HCP and HMW levels in adsorptive filterate. HCP and HMW content in filtrate samples for mAb A (HCP: 5000 ± 1000 ppm and HMW: 6.9 ± 0.6%) at a target loading of 200 L/m2. (A) Relative HCP levels were compared across filters as a function of increased loadings. (B) Relative HMW levels were compared were compared across filters as a function of increased loadings.
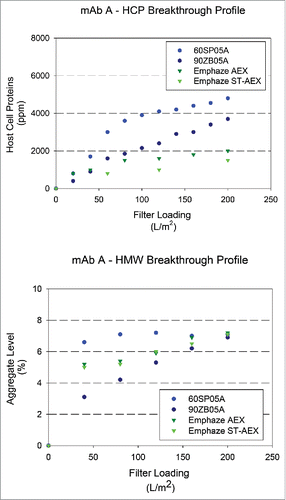
Figure 5. Breakthrough profiles for HCP and HMW levels in adsorptive filterate. HCP and HMW content in filtrate samples for mAb B (HCP: 2500 ± 400 ppm and HMW: 4.5 ± 0.2%) at a target loading of 200 L/m2. (A). Relative HCP levels were compared across filters as a function of increased loadings. (B). Relative HMW levels were compared were compared across filters as a function of increased loadings.
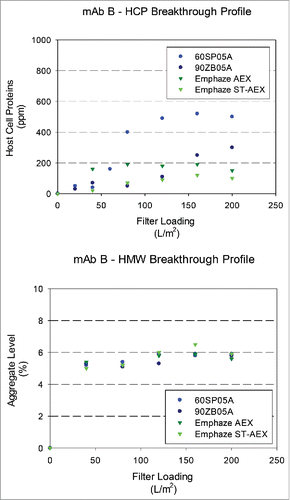
Figure 6. Breakthrough profiles for HCP and HMW levels in adsorptive filterate. HCP and HMW content in filtrate samples for mAb C (HCP: 28000 ± 8000 ppm and HMW: 9.1 ± 0.9%) at a target loading of 200 L/m2. (A). Relative HCP levels were compared across filters as a function of increased loadings. (B). Relative HMW levels were compared were compared across filters as a function of increased loadings.
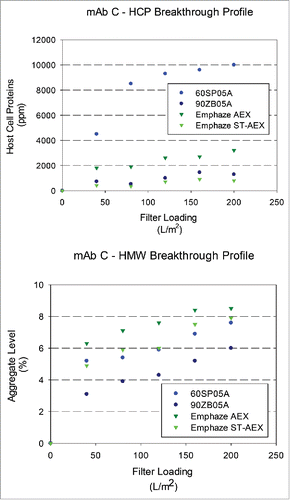
Figure 7. HCP content as a function of pH at a target loading of 200 L/m2. Host cell protein (HCP) content in filtrate samples remains lower at higher supernatant pH, which is consistent with an interaction between negatively charged HCP and positively charged filters. (A). Relative HCP levels were compared across runs for mAb A. (B). Relative HCP levels were compared across runs for mAb B.
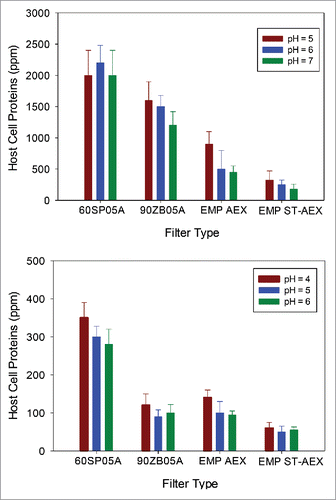
Figure 8. Reduced DNA levels as a function of pH at a target loading of 200 L/m2 for mAb A. DNA levels insensitive to higher supernatant pH, which is consistent with interaction of highly negatively negatively charged DNA and positively charged filter.
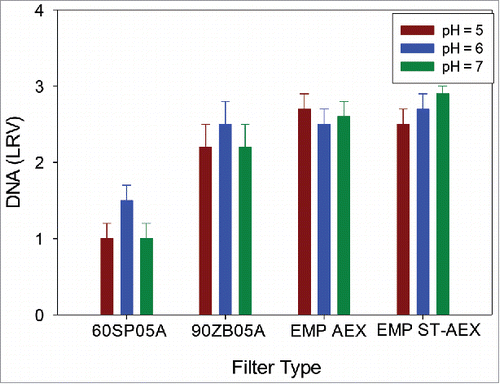
Figure 9. HCP levels in EMP ST-AEX filtrate pool as a function of salt concentration for mAb A and mAb B. HCP levels in EMP ST-AEX filtrate pools as a function of salt concentration at three different loadings (1, 3, and 5 kg/L). Data suggests improved HCP removal at higher salt concentration
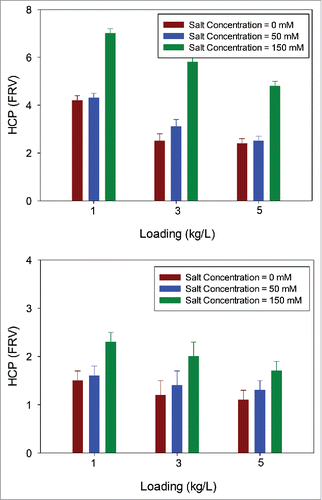
Figure 10. Aggregate Profiles of Protein A before and after ADF (90ZB05A) The size-exclusion chromatography (SEC) assay method quantifies the percent distribution of aggregates species (HMW1, HMW2, and HMW3) for mAb A and mAb C. 90 ZB05A shows removal most of HMW1 peak but not HMW2 and HMW3 species. HMW1 species showed higher hydrophobicity than HMW2 and HMW3 determined by hydrophobic interaction liquid chromatography (HIC-HPLC), suggesting the removal of HMW1 by ADF may be due to the hydrophobic interaction.
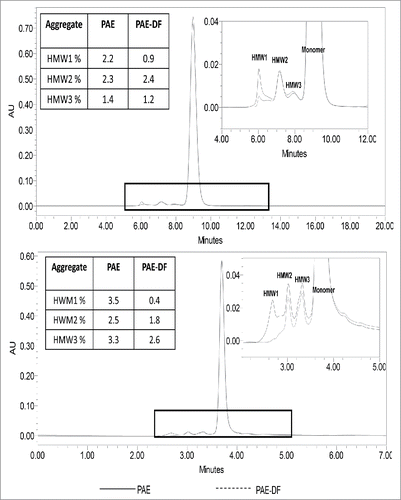
Table 3. PR772 and ΦX174 clearance in Post Protein A pools for three mAbs with adsorptive hybrid filters (AHFs).
Table 4. Comparison of conventional four-step purification of mAb A to the proposed two-step purification incorporating the adsorptive hybrid filter (AHF). Note that viral clearance data are not presented, but they are comparable in both these steps.
