Figures & data
Figure 1. Percentage of HIVAD8 Env gp140-specific memory (B) cells in total memory (B) cells. Antibody secreting cell number was calculated according to the number of spots formed on the plates coated with AD8 gp140. Cut-off spot forming unit was five per well. Data represents mean of 2 replicates (intra-variability) and error bars show standard deviation (SD). P values were calculated using one-way ANOVA followed by a Tukey honest significant difference (HSD) test post-test (ns, not significant; *P ≤ 0.05).
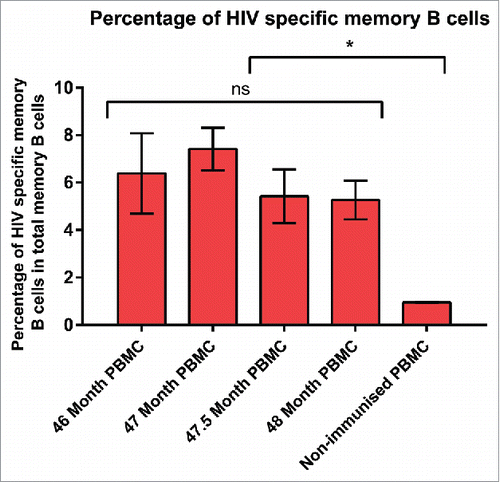
Figure 2. Gating strategy for gp140 binding IgG+ (B)cells from the vaccinated and unvaccinated cows. (A) Gating strategy for lymphocytes according to their size and granularity, (B) Gating on singlet cells, (C) Gating for viable CD21+ B cells, and (D) Gating of IgG+ CD21+ cells. Gp140 reactive memory IgG B cells (E) were identified as viable CD21+ IgG+ gp140+ cells within the lymphocyte gate.
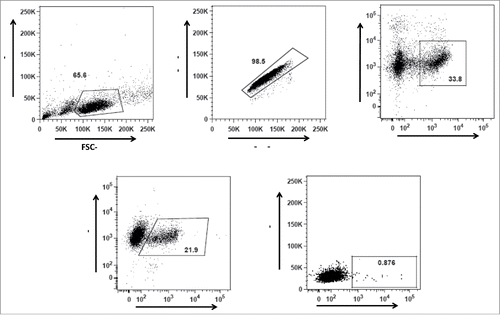
Figure 3. Bovine genes amplification and expression. (A) Representative of bovine heavy and light constant gene amplification. (B) Representative of amplification of Igγ and Igλ genes in single cell RT-PCR. The fragments amplified in the second round PCR were heavy Igγ (350–500bp) and light Igλ (320–340bp). (C) Representative cell culture supernatants of chimeric mAbs on a 12% reducing gel. Negative control includes the supernatant of cells mock transfected. The supernatant of VRC01 transfection was used as positive control for assessment of transfection and western blotting.
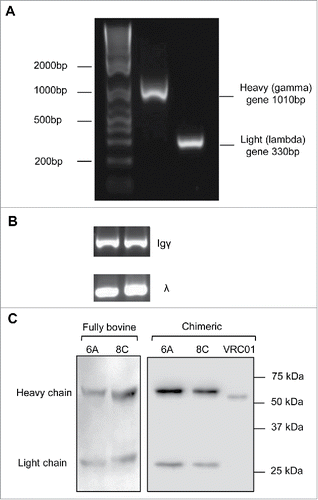
Figure 4. Env-binding of 6A and 8C chimeric mAbs against HIV Env. (A) HIV AD8 Env gp140 and (B) HIV AD8 Env. gp120 was immobilized on 96-well plates and serial dilutions of chimeric 6A and 8C mAbs were incubated with the coated wells. The binding of antibodies was detected with anti-human IgG-HRP conjugated.
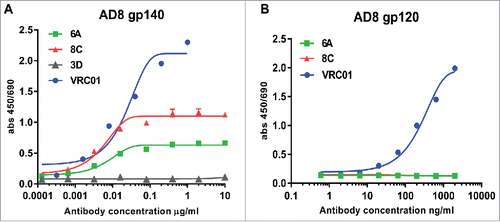
Figure 5. Binding of mAbs to the chimeric (HIV/SIV) AD8 gp140. (A) SIVgp41-AD8 gp120 IP assay: 6A/8C/VRC01-prt G controls (lanes: 2, 5, 8) were used to confirm the specific Env protein capturing in 6A/8C/VRC01-gp140-prt G samples (lanes: 3, 6, 9). gp41-AD8 gp120-prt G (lane: 10 show the non-specific background of chimeric gp140 binding to prt G in IP assay. 6A/8C/VRC01 controls (lanes: 1, 4, 7) included purified mAb in western blot assay to discriminate any possible western blot related background from real captured chimeric gp140 bands. Gp140 control (lane: 11) was used in western blotting assay to confirm the correct size of captured Env proteins in IP assay. (B) SIVgp120-AD8 gp41 IP assay; the control included: western blot control of 2F5, 6A and 8C (lanes 12, 15, 18), IP assay controls (13, 16, 19). The captured chimeric gp140 Env from 2F5, 6A and 8C IP assay could be detected in lanes 14, 17 and 20. Lane 21 represents the background on chimeric SIV gp120-AD8 gp41 non-specific binding to prt G. Lane 22 is the control samples which shows the position of SIV gp120-AD8 gp41.
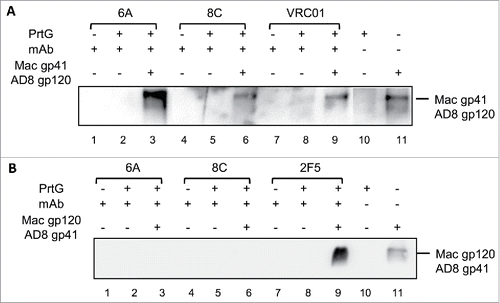
Figure 6. Binding of 6A and 8C mAbs to HIV AD8 Env. Binding to soluble trimeric (A) AD8 gp140 and (B) SOSIP gp140. 6A/8C/VRC01-prt G controls (lanes: 2, 5, 8) were used to confirm the specific Env protein capturing in 6A/8C/VRC01-soluble gp140/SOSIP gp140-prt G samples (lanes: 3, 6, 9). 6A/8C/VRC01 controls (lanes: 1, 4, 7) included 2µg of mAb (100ng for VRC01) in western blot assay. Soluble gp140/ SOSIP gp140 control (lane: 10) was used in western blot assay to confirm the correct size of captured Env proteins in IP assay. Lane 11 includes the sample with prt G only in IP assay.
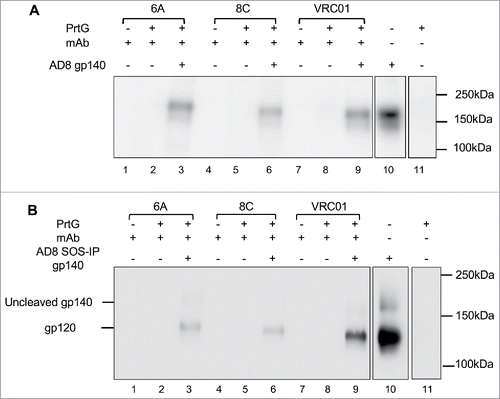
Figure 7. Competition of sCD4 with purified mAbs. Env AD8 gp140 was immobilized, then serial dilution of mAbs 6A, 8C and b12 with constant amount of 20 ug/ml sCD4 was added to each well. Antibody binding was detected as described in Material and Methods. Values represent the mean of two independent replicates.
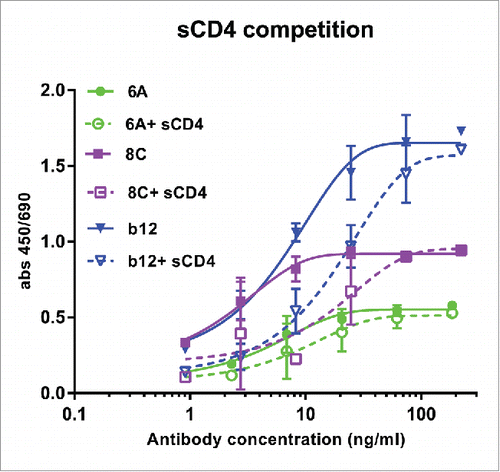
Figure 8. VH and CDRH3 size of selected AD8 gp140+ mAbs among isolated mAbs. (A) The length of VH region. Black points show all selected mAbs. Red point represents 6A mAb and blue point 8C mAb. (B) CDRH3 length of selected mAbs. The size of CDRH3 was defined as the number of aa in this region. Blue columns represent the number of all selected B cell clones while the black columns represent the number of residues in CDRH3 of 6A and 8C mAbs.
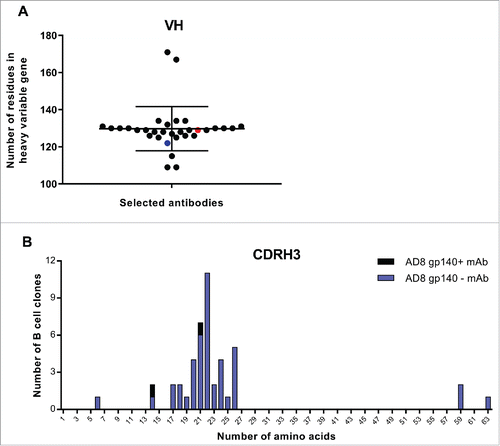
Figure 9. SHM and Cys and aromatic residues in VH and CDRH3 regions. (A) Number of Cys. (B) The average percentage of Cys in total CDRH3 residues. Black points show all selected mAbs. Red point represents 6A mAb and blue point shows 8C mAb. Green ovals represent the mAbs with the value above the average. (C) Number of aromatic residues. (D) The average percentage of aromatic residues in total CDRH3 residues. (E) Percentage of SHMs in CDRH3 of selected mAbs. (F) Percentage of SHMs in VH of selected mAbs. (G) 6A and 8C VH genes alignment with germline genes.
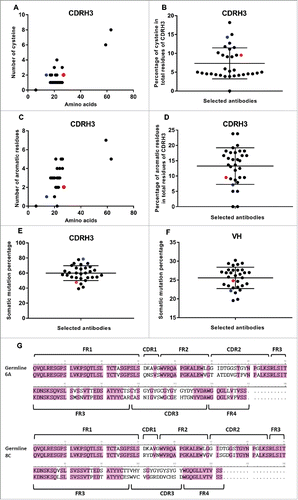
Table 1. Effect of mutations on structural change and Env-binding.
Figure 10. ELISA assay of mutated 6A and 8C mAbs. Ala mutagenesis of aromatic residues, His and Cys in heavy CDRs of 6A and 8C mAbs. (A-B) Soluble gp140-specific binding of wild type and mutated 6A and 8C mAbs against AD8 Env. Cut-off signal was 2 times above the background. Values represent the mean of three replicates.
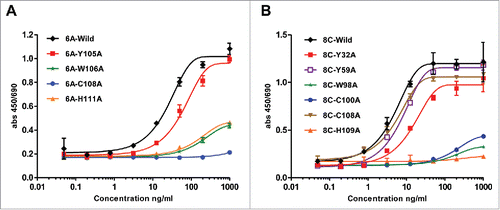
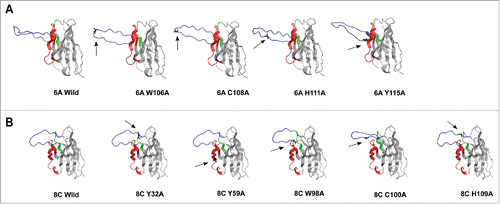
Figure 11. 3D structure homology-modeling of wild type and mutated 6A and 8C bovine V genes. (A) 6A wild type and mutated mAbs. (B) 8C wild type and mutated mAbs. The 8C and 6A VH regions are shown in gray and the heavy CDRs are as follow: CDRH1: green, CDRH2: red and CDRH3: blue. The mutated residues are shown as black and with arrow in each mutated mAbs. the VH region was modeled using a fully automated protein structure homology-modeling server (SWISS-MODEL, http://swissmodel.expasy.org). The server used bovine ultra-long CDRH3 as template (Protein data bank code: 4k3d.1.A for 6A mAb and 4k3e.2.A for 8C mAb). The figures were generated using PyMOL program.Citation88