Figures & data
Figure 1. Anti-GIPR antibodies raised in chicken vs. mouse and rat hybridoma antibodies. (A) illustrates the number and functional activity (antagonism as measured in GIPR specific cAMP assay) of anti-GIPR antibodies raised in chicken vs. previous campaigns using classic mouse and rat hybridoma technology. In (B), the chicken antibodies are broken down into those containing cysteines in the CDR3 of the heavy chain (Cys) and those without (no Cys) while also giving the fraction of antagonistic vs. not antagonistic antibodies in each population.
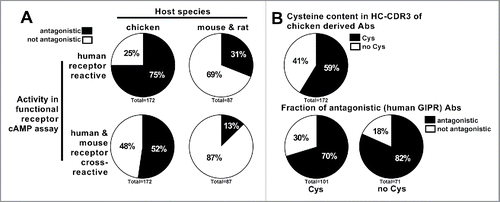
Table 1. Numerical representation of chicken-derived anti-GIPR antibodies.
Figure 2. Affinity data measured by surface plasmon resonance for anti-GIPR antibodies. (A) summarizes the SPR-derived KD values for anti-GIPR antibodies. The left side shows chicken-derived antibody affinities against human, mouse and rat-GIP receptor extracellular domain (ECD), whereas the right side provides human and mouse GIP receptor ECD affinities for the legacy mouse and rat hybridoma campaigns for comparison. All affinities are shown, irrespective of whether an antibody is antagonistic or not. (B) breaks down the KD values by the antagonistic activity of the chicken-derived antibodies in human or mouse receptor G specific functional cAMP assays. In (C), antibody KD values are grouped by the host chicken, whereas (D) lists chicken-derived antibody affinities based on whether or not they contain a cysteine in the heavy chain CDR3 (left side), as well as by functional activity against human and mouse GIPR and cysteine content (right side). The red lines indicate the population medians. P-values were calculated using Kruskal-Wallis and Mann-Whitney tests. ns - not significant; * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
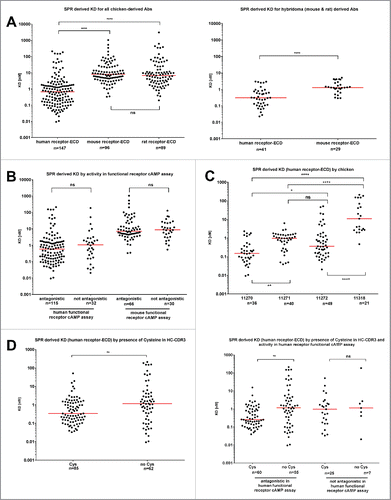
Figure 3. Functional activity of chicken-derived anti-GIPR antibodies in cAMP assay. IC50 values for the chicken-derived Abs were determined using an αscreen cAMP assay. (A) illustrates obtained values for functional antibodies broken down by cysteine content, whereas (B) illustrates antibodies raised in different chicken. (C) shows a correlation plot of the KD values vs. the IC50. In (D), the SPR off-rate constants (kd) are compared between antagonistic and not antagonistic chicken-derived antibodies. The red lines indicate the population medians. P-values were calculated using Kruskal-Wallis and Mann-Whitney tests. ns - not significant; * p<0.05; *** p<0.001.
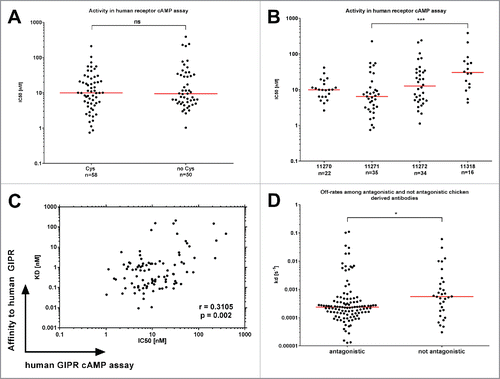
Figure 4. HC-CDR3 length of anti-GIPR chicken antibodies and effect on affinity and function. The histogram (A) lists the HC-CDR3 length distribution of cysteine-containing (black bars) vs. cysteine-free (gray bars) chicken-derived anti-GIPR antibodies. (B and C) list the affinities (KD) and antagonistic activities (IC50) among antibodies with short (< 19 amino acids) vs. long (≥ 19 amino acids) CDR3 sequences, respectively. The red lines indicate the population median. P-values were calculated using the Mann-Whitney test. ns - not significant; ** p<0.01.
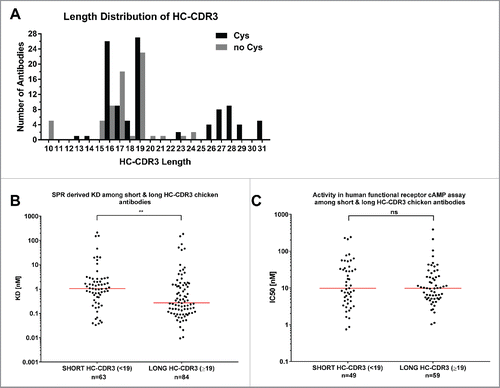
Figure 5. Biolayer interferometry-derived epitope clustering for anti-GIPR antibodies. A 2-dimensional matrix of the normalized biolayer interferometry assay data used for epitope clustering is shown. 45 anti-GIPR antibodies were assessed, 40 derived from chicken, 5 from other sources (rodent and phage display). The secondary antibodies are shown as rows, the primary antibodies as columns. Rows were sorted by their Pearson correlation coefficient (penultimate column on the right). Following Pearson row sorting, the columns were sorted to match the rows - hence the self-blocking value for each antibody is found on the diagonal (values marked in red). In addition, the Pearson correlation coefficient for the columns is shown in the bottom row. A color gradient from blue (0) to white (100) was applied to the data to highlight cross-blocking or competition. The last-most column indicates the epitope cluster an antibody was assigned based on the dendrogram shown in .
Figure 6. Dendrogram of BLI epitope clustering data. The dendrogram representing clustering of the secondary antibodies was generated in pvclust (see Materials and Methods). The axis on the left (Height) serves as a measure for antibody dissimilarity. Using a height cut-off of 10, 5 antibody clusters (A, B, C, D, R - red dashed boxes emerge). The AU value (%) represents the approximate unbiased p-value computed by pvclust using multiscale bootstrap resampling, whereas the BP value (%) indicates the bootstrap probability. Clusters with high AU values are strongly supported by the data. * denominates non-antagonistic antibodies.
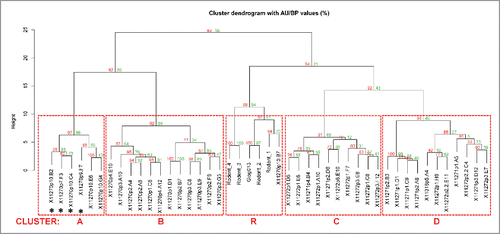
Figure 7. Phylogenetic tree of 40 chicken-derived anti-receptor G antibodies and cluster assignment. The phylogenetic tree (left panel) for the 40 chicken-derived anti-receptor G antibodies used for epitope clustering () was generated in MegAlign using ClustalW alignment of all full length VH sequences. Bootstrap percentage values are shown on each node. The table (right panel) lists antibody IDs, presence of cysteines, antagonistic activity as well as length and amino acid sequence of the HC-CDR3. The symbols in the penultimate column indicate antibodies with identical HC-CDR3s. The last column lists the BLI epitope cluster assignment derived from the dendrogram shown in .

Table 2. Chicken immunization regimes.
Table 3. Rodent immunization campaigns.
Table 4. SPR affinity and cAMP activity of anti-GIPR reference antibodies from rodent immunization and phage display.
