Figures & data
Figure 1. Design of the Fc tandem molecules. (A) Schematic representation of IgG1, IgG-2Fc and IgG-3Fc molecules. The IgG-2Fc contains 2 copies of Fc and has an expected molecular weight (MW) of 200 kDa, IgG-3Fc contains 3 copies of Fc and has an expected MW of 250 kDa. (B) The connecting region between Fcs include 2 (GGGGS) repeats and a full hinge region. (C) Schematic representation of the alternatively folded IgG-2Fc-half molecule with a MW of 100 kDa.
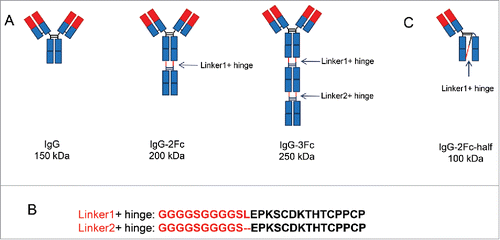
Figure 2. Expression and purification of IgG tandem Fc variants. (A) Mammalian cell-expressed IgG1, IgG-2Fc and IgG-3Fc proteins from 2 clones, anti-LPS 6F6 and anti-MrA KP3, were captured by protein A beads and analyzed by SDS-PAGE under reducing and non-reducing conditions. (B) The IgG-2Fc-half, IgG-2Fc and IgG-3Fc fractions were separated by an additional ion-exchange chromatography step and further purified. The purity and size of each IgG Fc variants were confirmed by a SDS-PAGE analysis. M, molecular weight (MW) marker. Numbers on the left indicate the MW in kDa. “*” denotes the location of the IgG-2Fc-half molecules under non-reducing condition.
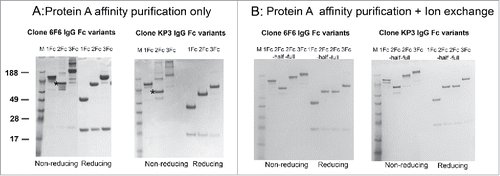
Figure 3. Binding profiles of IgG Fc variants to Fc receptors, C1q and antigens. (A) ELISA evaluation of 6F6 IgG Fc variants binding to Fc receptors including FcγRI, FcγRIIa, FcγRIIIa(158F), FcγRIIb,FcRn and C1q. (B) ELISA evaluation of binding activity of anti-MrkA KP3 and anti-LPS 6F6 IgG Fc variants to their respective antigens.
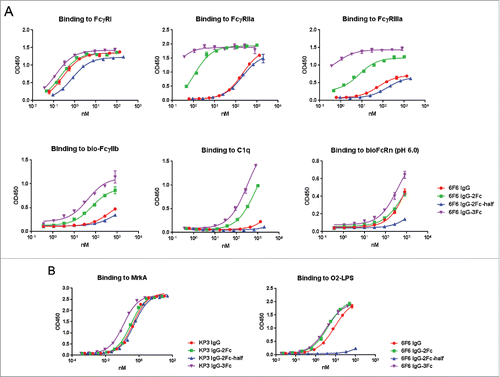
Figure 4. Size-exclusion high performance liquid chromatography (SE-HPLC) analysis of the purity of monomeric IgG Fc variants after prolonged storage. 6F6 (upper panels) and KP3 (lower panels) IgG variants including IgG1, IgG-2Fc-half, IgG-2Fc and IgG-3Fc were stored at 4°C for one year and analyzed by a SE-HPLC. Monomer percentage of each of the Fc variants is shown in each graph. Monomer percentages experiencing significant decrease (> 3%) are highlighted with a red box.
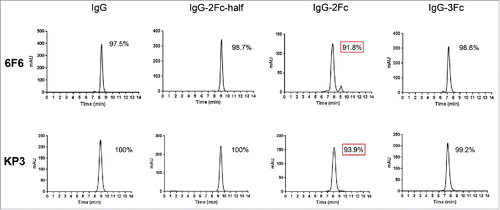
Figure 5. Differential scanning calorimetry (DSC) analysis of the thermal stability of 6F6 IgG Fc variants. DSC profiles of IgG1 (A), IgG-2Fc-half (B), IgG-2Fc (C) and IgG-3Fc (D) are presented.
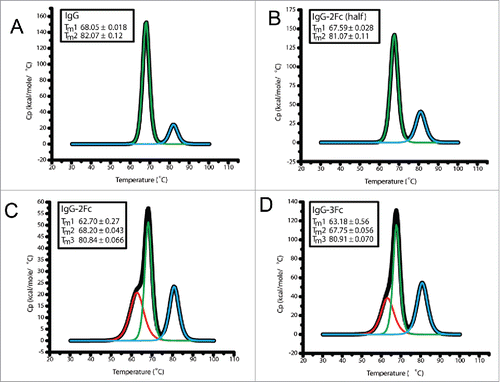
Figure 6. Pharmacokinetic profiles of KP3 IgG Fc variants in C57BL/6 mice. The serum levels for each of the IgG variants were determined by an ELISA. Each data point represents the average IgG concentration in sera from 3 mice for different time points up to 7 d.
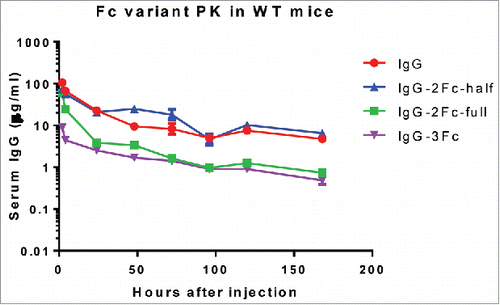
Table 1. Pharmacokinetic parameters of KP3 Fc variants following 10 mg/kg IV in mice. ND: not determined because of data fluctuation; Cmax = maximal observed concentration; AUClast = area under the concentration time curve up to the last measurable concentration; AUC∞ = Area under the curve from time zero to infinity; T1/2 = terminal phase elimination half- life; CL = systemic clearance; IV = intravenous injection.
Figure 7. Opsonophagocytic killing (OPK) activity of IgG Fc variants against different Klebsiella pneumoniae (KP) strains. The OKP activity of 6F6 IgG variants were tested against KP 5053 (A) and 8570 (B) strains. The activity of KP3 IgG variants were tested against KP 29011 (C) and 961842 strains (D). The percentage of killing was determined for different IgG Fc variants against an isotype control IgG.
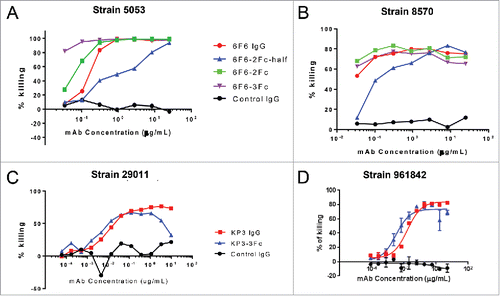
Figure 8. In vivo protective activity of IgG Fc variants. The in vivo activities of KP3 IgG Fc variants (A) and 6F6 IgG Fc variants (B) were evaluated in acute pneumonia mouse models. C57BL/6 mice were inoculated with a multi-drug resistant isolate intra-nasally. IgG Fc variants were administered one hour post bacterial infection. Mouse survival was monitored daily up to day 7 post challenge. For the evaluation of the KP3 variants, 5.22×107 CFU of KP strain 29011 were inoculated intra-nasally to the mice one hour before the IV injections of the KP3 Fc variants. For the evaluation of the 6F6 variants, 1.32×104 CFU of KP strain 3048570 were inoculated intra-nasally to the mice one hour before the IV injections of the 6F6 Fc variants. R347 is a human IgG1 isotype (negative) control. A rabbit polyclonal antibody generated against a wild type KP strain was included as a positive control. There were 10 animals in each group in (A) and 8 animals in each group in (B).
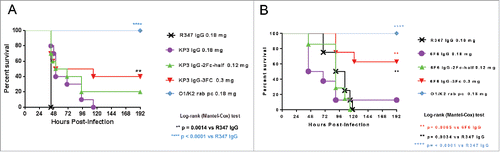
Figure 9. Binding profiles of IgG Fc variants with Fc point mutations to FcγRs, C1q and antigens. The 2 isoforms of FcγRIIIa (158F and 158V) are indicated.
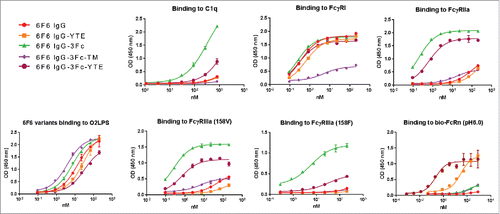
Figure 10. Pharmacokinetic profiles of 6F6 IgG Fc variants in human FcRn transgenic mice. (A) The serum levels for each of the IgG variants were determined by an ELISA. Each data point represents the average IgG concentrations in sera from 3 mice for up to 9 d. (B) 3Fc-YTE induced significant anti-antibody response in mice whereas 3Fc did not. In a direct ELISA, 6F6–3Fc and 6F6–3Fc-YTE were coated to an ELISA plate directly and subjected to detection with sera from mice injected with 6F6–3Fc or 6F6–3Fc-YTE, respectively. The sera of representative mice from both groups collected at day 4 (upper panel) and day 9 (lower panel) were used in the ELISA.
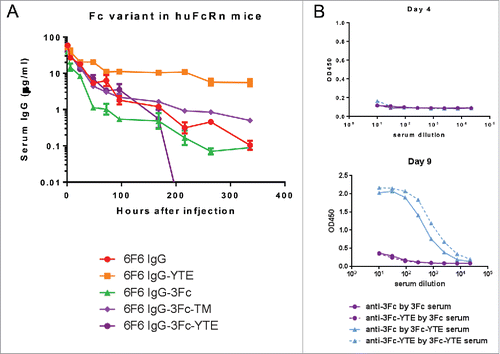
Table 2. Pharmacokinetic parameters of KP3 Fc mutant variants following 2 mg/kg IV in huFcRn transgenic mice. The parameter symbols used are the same as in .
