Figures & data
Figure 1. The antigen-contacting residues on the structures of 5 RabMAbs in the PDB. An antigen-contacting residue (highlighted in red) contains at least one atom that is ≤ 6 Å away from an atom on the antigen (not shown). The approximate locations of CDRs are labeled. Aligned: RabMAbs are structurally aligned with 2×7L by Dali pairwise comparison and visualized by Chimera.
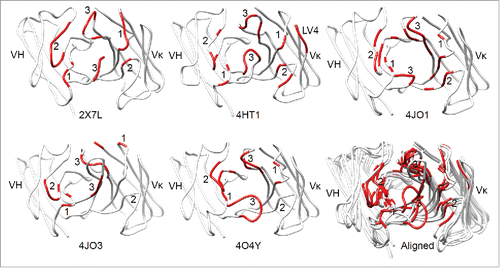
Figure 2. Multiple structure alignment of RabMAb VHs by Strap. Antigen-contacting residues, their supporting residues, and Kabat/IMGT/Paratome CDRs are indicated in protein sequences. HV4, a non-CDR loop in heavy chain FR3.
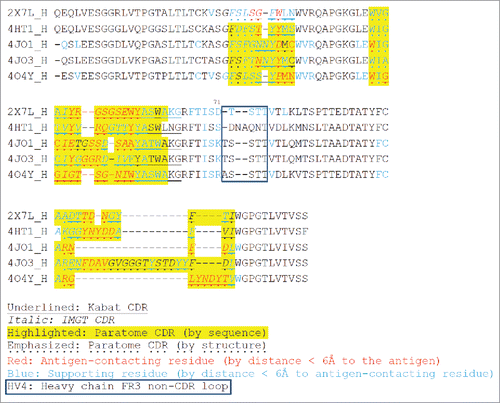
Figure 3. Multiple structure alignment of RabMAb Vκs by Strap. Antigen-contacting residues, their supporting residues, and Kabat/IMGT/Paratome CDRs are indicated in protein sequences. LV4: a non-CDR loop in light chain FR3.
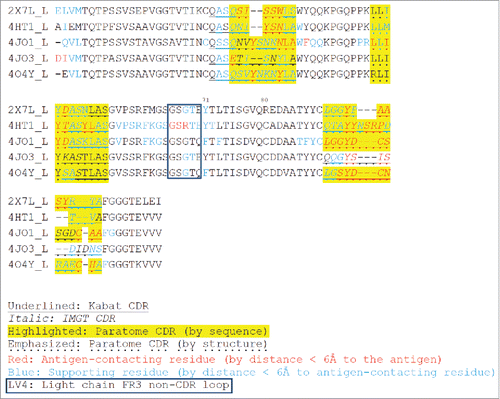
Table 1. Identification of the most similar rabbit germline sequences for the 5 rabbit Fvs in the PDB database by IMGT/DomainGapAlign.
Table 2. The number of antigen-contacting residues in CDRs.
Figure 4. Humanization of RabMAbs via grafting combined Kabat/IMGT/Paratome CDRs. The YP218 RabMAb and its humanized antibody (hYP218) structures were modeled by I-TASSER, aligned by Dali pairwise comparison, and visualized by Chimera.
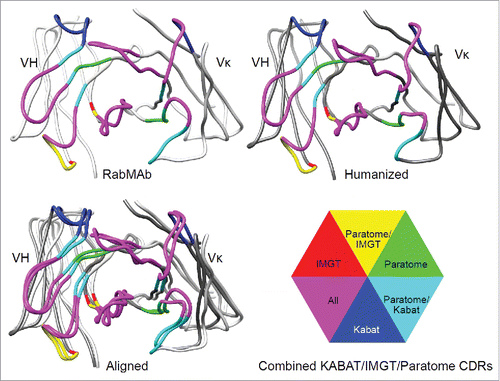
Figure 5. The sequence alignment of rabbit Fv and human germline sequences used as templates. Human germline IGHV3–66*01, IGHJ4*01, IGKV1–27*01 and IGKJ4*01 were used as the templates for humanization. FW1.4 and FW1.4 gen were used as the templates in a previous RabMAb humanization report.Citation33. The additional cysteine in position 80 (Kabat numbering) on the rabbit Vκ is replaced with proline in human germline template (IGKV1–27*01).

Figure 6. The characterization of immunotoxins with humanized scFvs and original RabMAb scFvs. (a) Schematic cartoon of a monovalent scFv-based immunotoxin. (b) SDS-PAGE of purified immunotoxins. (c) Size-exclusion chromatogram of purified proteins. T: theoretical molecular weight calculated from the protein sequence. E: experimental molecular weight calculated by the MALS. (d) Association and dissociation curve on purified proteins, measured on the Octet platform. *: for hYP223IT, 282.6 nM instead of 300 nM immunotoxin protein was used. (e) Immunotoxin binding curve on H9 cells by flow cytometry. BL22 is an irrelevant immunotoxin (anti-CD22) used as a negative control.
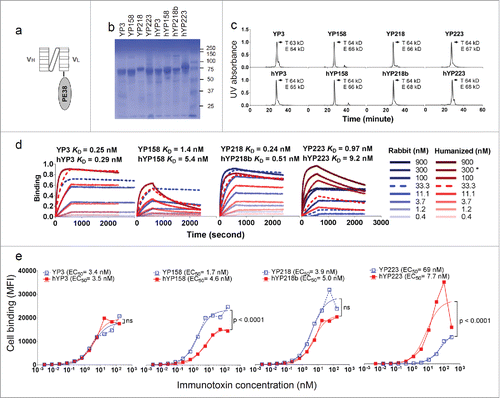
Figure 7. The cytotoxicity induced by immunotoxins with humanized scFvs and original RabMAb scFvs. They were tested on H9 cells in a WST assay. Error bars indicate standard errors. YP7, an irrelevant immunotoxin to GPC3.
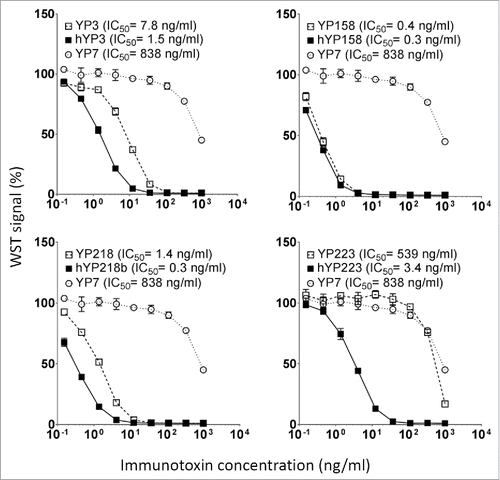
Table 3. The effect of position 71 on RabMAb humanizations.
Table 4. The binding kinetics of purified immunotoxins containing rabbit or humanized scFv. The binding kinetics were measured by Octet and calculated by Graphpad Prism. The cell surface binding EC50 on H9 cells are listed for comparison.
