Figures & data
Figure 1. Molecular model of MP0250 consisting of four designed ankyrin repeat domains in a single polypeptide chain. The target specificity of every domain is indicated in the figure. For every designed ankyrin repeat domain, positions that were randomized in the DARPin® library and thus represent potential target interaction residues, are colored yellow (serum albumin), red (HGF), or orange (VEGF).
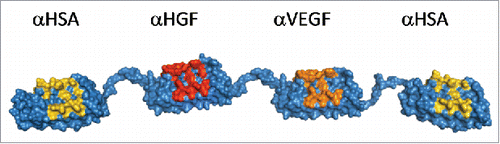
Table 1. Shake-flask expression levels of MP0250 and DARPin® domains (E. coli BL21, TB medium).
Figure 2. Storage stability of MP0250 assessed by reverse-phase HPLC analysis. MP0250 at ∼ 15 mg/mL in PBS was stored for up to 24 months at −70°C (open circles), −20°C (open squares), 5°C (open triangles), or 25°C (open diamonds) in glass vials. Reverse-phase HPLC (see Materials & Methods) was used to determine the % of main peak at 1, 2, 3, 6, 9, 12, 18, or 24 months of incubation. In frozen state at −70°C and −20°C, the % main peak value in reverse-phase HPLC is stable with only minor change. Similarly, the % main peak value for MP0250 stored at +5°C is stable with only minor change over the time measured. As expected, a decrease in % of main peak was observed when MP0250 was incubated at +25°C for several months.
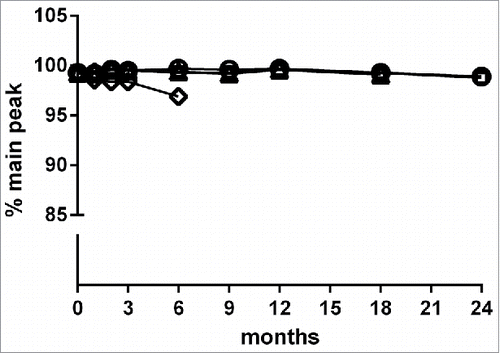
Figure 3. Potent binding of MP0250 in ELISA. (a) VEGF-A binding ELISA. The binding signal of various concentrations of MP0250 to immobilized VEGF-A of human (filled circles; 100% sequence identity to cynomolgus monkey VEGF-A), and mouse (filled diamonds), as well as human VEGF-C (open inverse triangles), and human PDGF-AB (open circles), and the corresponding fitting inhibition curves are shown. (b) HGF binding ELISA. The binding signal of various concentrations of MP0250 to immobilized HGF of human (filled circles), cynomolgus monkey (filled triangles), and mouse (filled diamonds), and the corresponding fitting inhibition curves are shown. (c) Serum binding ELISA. The binding signal of various concentrations of MP0250 to immobilized serum of human (filled circles), cynomolgus monkey (filled triangles), and mouse (filled rhombus) and the corresponding fitting inhibition curves are shown. For all graphs, the ELISA signal (OD 450 nm – OD 620 nm) is plotted as a function of the MP0250 concentration in pM. Experimental details are described in the Materials & Methods section. Fitted EC50 values are shown in .
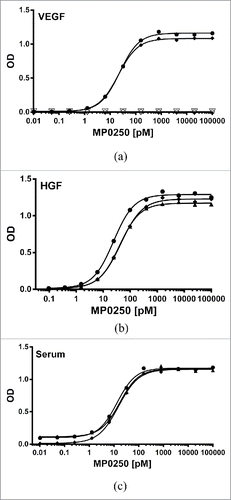
Table 2. Apparent EC50 values (and 95% confidence interval) of MP0250 for binding VEGF-A, HGF and serum albumin of different species.
Figure 4. Comparison of His-tagged MP0250 (circles) in comparison to individual DARPin® domains (triangles) in cellular assays. (a) HUVEC proliferation inhibition assay. MP0250 is equivalently potent as the individual VEGF-inhibiting DARPin® domain of MP0250 in inhibiting HUVEC proliferation. Reference (square): sample with no inhibitor present. (b) c-Met phosphorylation inhibition assay. MP0250 is equivalently potent as the individual HGF-inhibiting DARPin® domain of MP0250 in inhibiting c-Met phosphorylation. The cellular assays were performed as described in the Materials & Methods section.
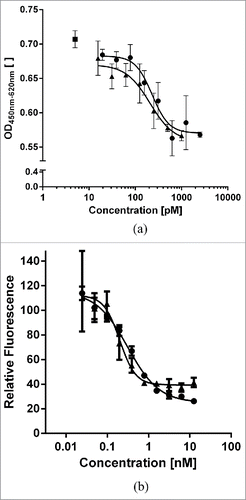
Figure 5. Surface plasmon resonance measurement to show simultaneous binding of MP0250 to HGF, VEGF-A, and human serum albumin. Setup: Human HGF is immobilized, MP0250 is injected (time 0 to 180 seconds) followed by a short buffer injection, human VEGF-A is then injected (time 600 to 780 seconds) followed by a short buffer injection, and finally HSA is injected (time 1200 to 1380 seconds), followed by a longer buffer injection. Trace 1 is the full measurement, traces 2–6 are control measurements (2: No HSA injected; 3: no human VEGF-A injected; 4: No MP0250, no human VEGF-A, and no HSA injected; 5: No MP0250 and no HSA injected; 6: No MP0250 and no human VEGF injected). RU: resonance units [ ]; t: time [s]. The measurement indicates that MP0250 binds to the immobilized HGF and can then bind VEGF-A to full saturation, indicating complete HGF/VEGF/MP0250 complex formation. This complex is then able to bind HSA, indicating that MP0250 can bind all targets simultaneously. The maximum signal of trace 1 being higher than trace 3 indicates that at least a fraction of the complexes consists of the full MP0250/HGF/VEGF-A/HSA complex. The controls indicate that no unspecific binding occurs in the measurement.
![Figure 5. Surface plasmon resonance measurement to show simultaneous binding of MP0250 to HGF, VEGF-A, and human serum albumin. Setup: Human HGF is immobilized, MP0250 is injected (time 0 to 180 seconds) followed by a short buffer injection, human VEGF-A is then injected (time 600 to 780 seconds) followed by a short buffer injection, and finally HSA is injected (time 1200 to 1380 seconds), followed by a longer buffer injection. Trace 1 is the full measurement, traces 2–6 are control measurements (2: No HSA injected; 3: no human VEGF-A injected; 4: No MP0250, no human VEGF-A, and no HSA injected; 5: No MP0250 and no HSA injected; 6: No MP0250 and no human VEGF injected). RU: resonance units [ ]; t: time [s]. The measurement indicates that MP0250 binds to the immobilized HGF and can then bind VEGF-A to full saturation, indicating complete HGF/VEGF/MP0250 complex formation. This complex is then able to bind HSA, indicating that MP0250 can bind all targets simultaneously. The maximum signal of trace 1 being higher than trace 3 indicates that at least a fraction of the complexes consists of the full MP0250/HGF/VEGF-A/HSA complex. The controls indicate that no unspecific binding occurs in the measurement.](/cms/asset/5efc358b-f87e-4f67-8d40-192e41cf7cde/kmab_a_1305529_f0005_b.gif)
Figure 6. Concentration-time profiles of MP0250. (a) Mouse pharmacokinetic profile of MP0250 at 1 mg/kg (filled circles) and 2 mg/kg (filled squares). (b) Cynomolgus monkey pharmacokinetic profile of MP0250 at 1 mg/kg (open circles), 10 mg/kg (open squares), and 100 mg/kg (open triangles). Mean concentrations and standard deviations are plotted over time. Pharmacokinetic parameters derived from the experiments are given in .
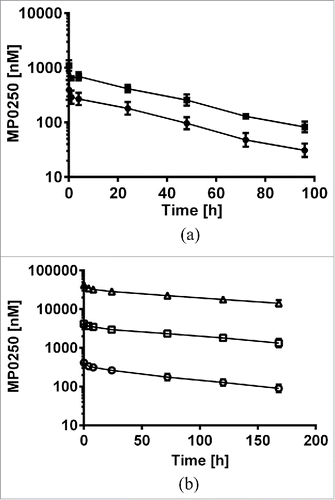
Table 3. Pharmacokinetic properties* of MP0250 i.v. administered in mouse and cynomolgus monkey.
