Figures & data
Figure 1. A schematic of GMD-FACS antibody screening. (A) P. pastoris and mammalian target cells are co-encapsulated in GMDs. During induction, P. pastoris secretes full-length mAb, which diffuses throughout the GMD matrix. (B) Secreted full-length mAbs can bind antigen targets on the surface of antigen-positive mammalian cells (lower) but not negative cells (upper), which lack the antigen. (C) Unbound antibody is removed from the GMDs by washing, and fluorophore conjugated secondary antibodies are added to selectively detect antigen-positive target cells (lower).
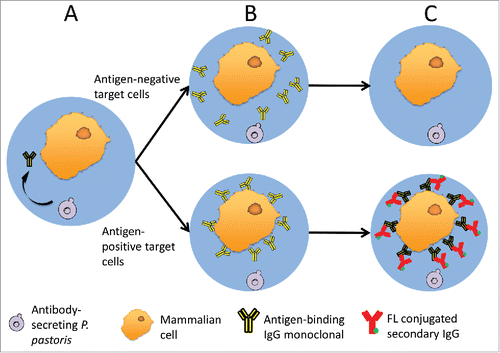
Table 1. Protein sequences and estimated affinities of anti-EGFR mAb and anti-CCR5 mAb.
Figure 2. Preliminary FACS analysis of antibody staining in GMDs. (A) A431 cells were encapsulated in GMDs and labeled with purified positive control anti-EGFR (red) and negative control anti-CCR5 (blue) antibodies, respectively. Empty GMDs (grey) were treated in an analogous fashion. FACS histograms following staining with secondary antibody are shown. (B) A431 cells were co-encapsulated with yeast expressing either anti-EGFR mAb (positive control, red) or anti-CCR5 mAb (negative control, blue), respectively. FACS histograms following staining with secondary antibody are shown. Results are representative of three independent experiments.
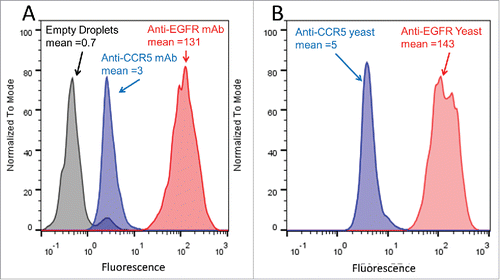
Figure 3. FACS sorting of GMD populations. (A) FACS dot plot showing forward scatter (FSC) and back scatter (BSC) signals of GMD samples. The left and bottom populations in gate R1 were a mixture of free A431 cells, GMD debris, and free P. pastoris cells. The right top population in rectangular gate R2 was intact GMDs that were either empty or contained A431 cells, P. pastoris cells, or both. Subsequent FACS sorts were gated on the R2 GMD population. (B) Micrograph image of representative GMDs before sorting. (C) PE signal of the GMDs from gate R2. Three fluorescence gates were used to sort GMDs based on PE signal intensity: the lowest 10% (R3), the central 80% (R4), and the highest 10% (R5). (D) Image of representative GMDs from the R3 sort gate. Most GMDs were empty or contained yeast cells only. (E) Image of representative GMDs from the R4 sort gate. Samples contained a large proportion of GMDs with only A431 cells. (F) Image of representative GMDs from the R5 sort gate. Samples contained a large proportion of GMD with both yeast and A431 cells. Scale bars are 50 µm. Results are representative of three independent experiments.
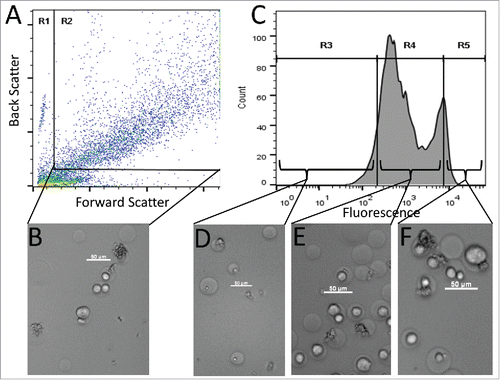
Figure 4. GMD-FACS signal as a function of antigen expression level and antibody affinity. (A) Yeast expressing anti-EGFR mAb were co-encapsulated with HT29 cells (low EGFR expression, black), A549 cells (moderate EGFR expression, blue), and A431 cells (high EGFR expression, red), respectively. Following an 18 hour mAb induction, staining with secondary antibody-PE conjugate, and FACS analysis, the fluorescence intensities (mean values provided) of the three GMD preparations correlated well with target cell EGFR expression level (approximate EGFR copy per cell provided as “N”). Yeast producing an anti-CCR5 antibody co-encapsulated with A431 cells (green) are shown as a negative control. (B) A431 cells, expressing high levels of EGFR, were co-encapsulated with yeast expressing anti-EGFR antibodies of different affinities: Nimotuzumab (low affinity, black), anti-EGFR mAb (moderate affinity, blue), and P2X (high affinity, red). Following a 12 hour mAb induction, staining with secondary antibody-PE conjugate, and FACS analysis, the fluorescence intensities of the three GMD preparations correlated well with antibody affinity to the EGFR target (approximate values provided in nM). Yeast producing an anti-CCR5 antibody (green) are shown as a negative control. Results are representative of two independent experiments.
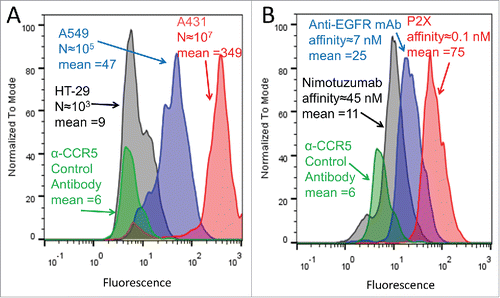
Figure 5. FACS sorting of a mock antibody library population. Positive control yeast (expressing anti-EGFR mAb) were mixed at a 1:10,000 ratio with negative control yeast (expressing anti-CCR5 mAb) followed by co-encapsulation with A431 target cells. (A) Schematic of library sorting strategy. (B) Population composition after each sort. Solid bars indicate the proportion of negative control yeast and checkered bars indicate the proportion of positive control yeast. Fold enrichment [(% anti-EGFR post sort)/(% anti-EGFR presort)] is shown.
![Figure 5. FACS sorting of a mock antibody library population. Positive control yeast (expressing anti-EGFR mAb) were mixed at a 1:10,000 ratio with negative control yeast (expressing anti-CCR5 mAb) followed by co-encapsulation with A431 target cells. (A) Schematic of library sorting strategy. (B) Population composition after each sort. Solid bars indicate the proportion of negative control yeast and checkered bars indicate the proportion of positive control yeast. Fold enrichment [(% anti-EGFR post sort)/(% anti-EGFR presort)] is shown.](/cms/asset/9b644d9b-345c-47a0-9d45-e774f73ebfea/kmab_a_1381812_f0005_oc.gif)
