Figures & data
Table 1. Binding and neutralization properties of isolated mAbs.
Figure 1. Epitope mapping of isolated mAbs. (a) Epitope binning with RSV PreF antigen. The heatmap shows epitope binning from the sandwich-based BLI binding assay, with darker green color and small number indicating more competition between the 1st and the 2nd antibodies. Gray sidebar on the left shows mAbs with PreF-preferred binding, determined by ELISA (without dashed box) or BLI (in dashed box); colored side-bar on the left shows mAbs of which epitopes were confirmed by site knock-out mutants. (b) Epitope binning with hMPV WT trimer antigen. Similarly, the heatmap shows epitope binning from the sandwich-based BLI binding assay, with darker green color and small number indicating more competition between the 1st and the 2nd antibodies.
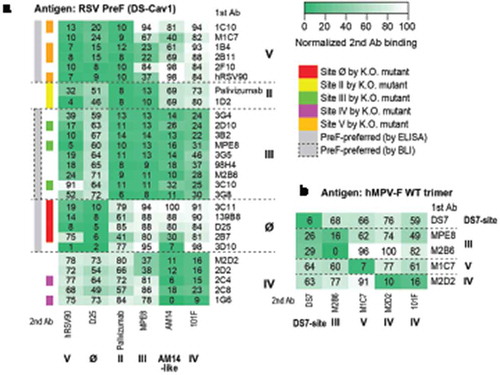
Table 2. V gene germline usage and somatic hypermutation analysis of isolated site III mAbs.
Figure 2. Characterization of site III mAbs with different binding and neutralization modes to hMPV. (a, b) ELISA binding of site III mAbs to wild-type hMPV-B F antigen and the A314N mutant. Error bars represent the standard deviation of experiment duplicates. (c, d) Critical residues for binding of 3G4, 98H4, and 3G8 mAbs to RSV PreF antigen, determined by alanine scanning library. For each experiment, HEK293T cells expressing the RSV F mutation library were tested for immunoreactivity with mAb of interest, measured on an Intellicyt high-throughput flow cytometer. Clones with reactivity of <30% relative to that of wildtype RSV F (horizontal line) yet >70% reactivity for a control mAb (vertical line) were initially identified to be critical residues for mAb binding (red dots), and were verified using algorithms described elsewhere.Citation52 The mean binding reactivities (and ranges) are shown in the tables. The critical residues are visualized on RSV PreF structure.Citation28.
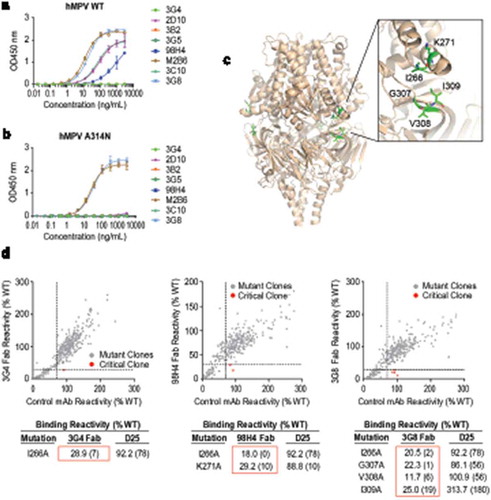
Figure 3. Characterization of two RSV/hMPV cross-neutralizing mAbs targeting site IV and V. (a) ELISA binding of M2D2 and M1C7 to wild-type hMPV-B2 F antigen. (b) Binding of M2D2 and M1C7 on RSV A PreF, RSV B PreF, and RSV B PostF antigens, determined by ELISA with Expi293 cell culture supernatants containing expressed antigens. (c) Neutralization assay of M2D2 and M1C7 to RSV A Long and B Washington strains. (d) Neutralization assay of M2D2 and M1C7 to hMPV A and B subtypes.
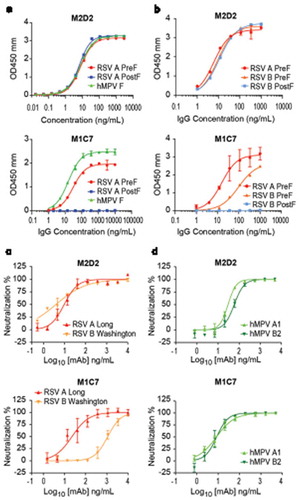
Figure 4. Epitope mapping of M1C7. (a) Binding of M1C7 on RSV A PreF antigen and derived K201N, S169N mutants, determined by ELISA with Expi293 cell culture supernatants containing expressed antigens. Error bars represent the standard deviation of experiment duplicates. (b) Critical residues for M1C7 binding, determined by alanine scanning library. For each experiment, HEK293T cells expressing the RSV F mutation library were tested for immunoreactivity with mAb of interest, measured on an Intellicyt high-throughput flow cytometer. Clones with reactivity of <10% relative to that of wildtype RSV F (horizontal line) yet >70% reactivity for a control mAb (vertical line) for M1C7 were initially identified to be critical residues for mAb binding (red dots), and were verified using algorithms described elsewhere.Citation52 The mean binding reactivities (and ranges) are shown in the tables. (c) The critical residues are visualized on RSV PreF structure.Citation28 (d) Sequence alignment of F antigens in hMPV A2, B2, RSV-A Long, RSV-B Washington strains near the epitope of M1C7. Numbers above sequences indicate the original residue number in the full-length sequence. Arrows indicate the five residues critical for M1C7 binding. The residues were colored in Crystal-X style by MOE.
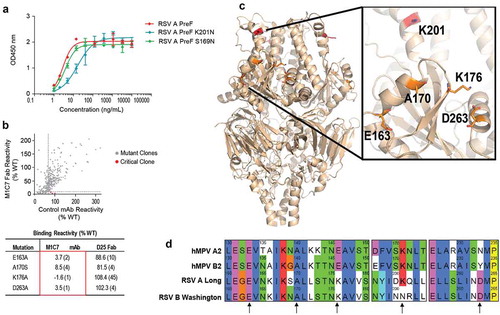
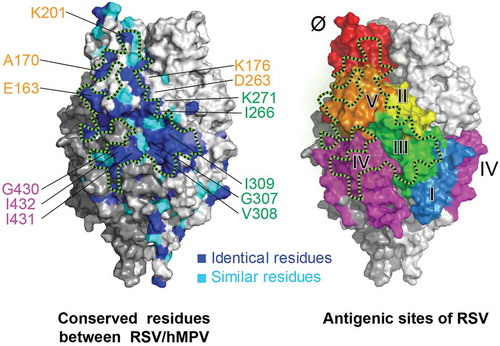
Figure 5. A conserved surface patch on F protein for RSV/hMPV cross-neutralization. Crystal structure of RSV PreF,Citation28 colored by conserved residues (blue and cyan) between RSV and hMPV (left) and antigenic sites (right). Black dashed lines circle a conserved surface patch between RSV/hMPV F antigens. Critical residues identified for antibody binding were labeled and color-coded according to the corresponding antigenic sites.