Figures & data
Figure 1. Analysis of CD160 library diversity after phage display enrichment
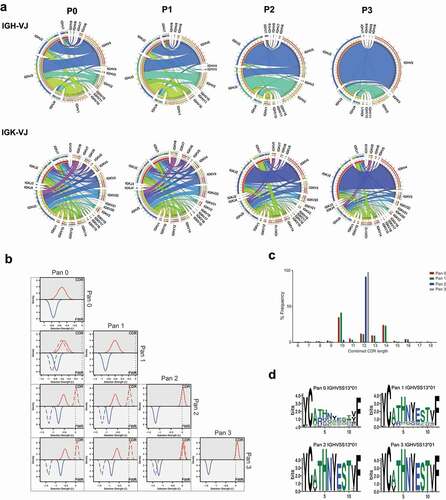
Figure 2. Heavy and light chain pairing in phage libraries
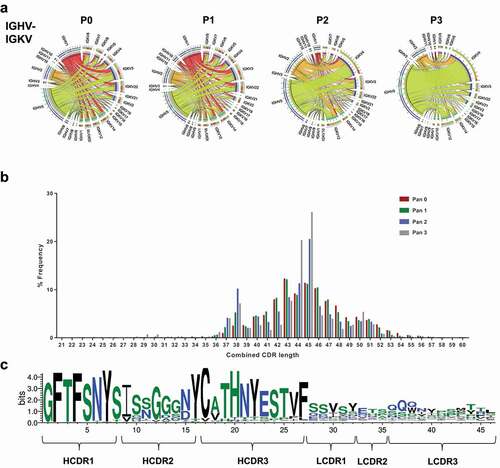
Figure 3. Overview of strategy to generate antibodies from deep-sequenced scFv libraries
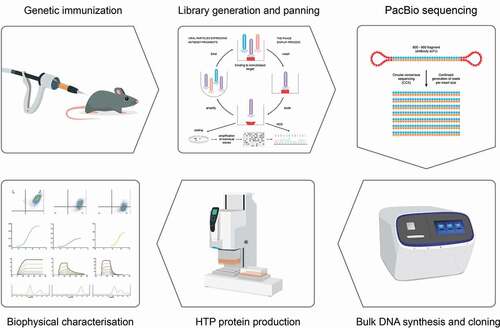
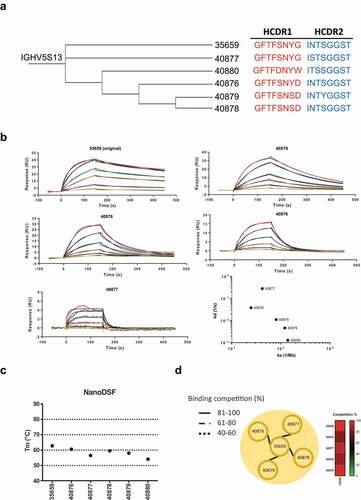
Figure 4. Identification and characterization of scFv clones
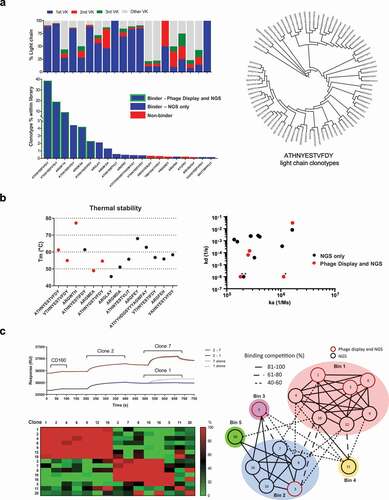
Table 1. Summary of the top 20 CD160 scFv identified
Figure 5. Identification of alternative binders in the NGS data set