Figures & data
Table 1. Number of positive and negative comparisons in the datasets, based on Ab-Ligity’s definition of similar epitopes
Table 2. Precision and recall on the full and CDRH3 ≤ 0.8 sets, using the selected paratope similarity thresholds for Ab-Ligity (0.1) and InterComp (0.6), based on Ab-Ligity’s definition of similar epitopes
Table 3. Precision and recall on the full set, using heavy chain and light chain only homology models and the predicted paratopes on the corresponding single domain
Figure 1. Analysis of anti-lysozyme (HEL) antibodies with dissimilar CDRH3 sequences and highly similar epitopes
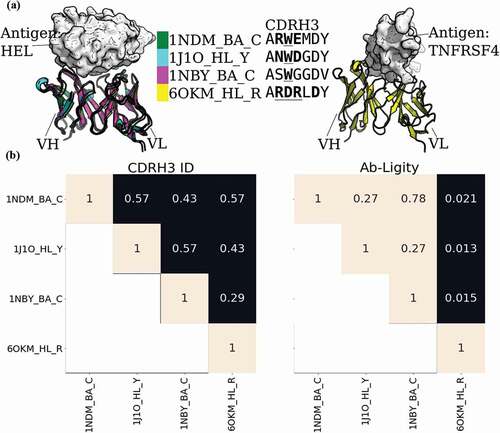
Figure 2. Analysis of two anti-gp120 antibodies. VRC01 antibody (PDB code and chain IDs: 4LSS_HL_G) is colored in blue; VRC07 (PDB code and chain IDs: 4OLU_HL_G) is shown in pink. The gp120 core antigen is displayed as white surfaces and the superimposed antibodies are in cartoons. The CDRH3 loops are in solid shades of the cartoon representation. The PDB code, heavy and light chain ID, and antigen chain ID are separated by (‘_’) and listed in the legend with the corresponding CDRH3 sequences. CDRH3 sequences are shown by aligning their IMGT positions and ‘-’ indicates a gap in the alignment according to the IMGT numbering scheme.Citation29 Crystal paratope residues within the CDRs are in bold, and Parapred-predicted paratopes within the CDRs are underlined. The other five CDR sequences are listed in Supplementary Table S16. The Ab-Ligity paratope similarity score for the pair is listed
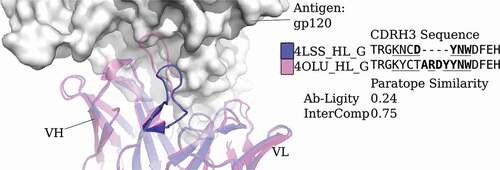
Figure 3. The Ab-Ligity workflow
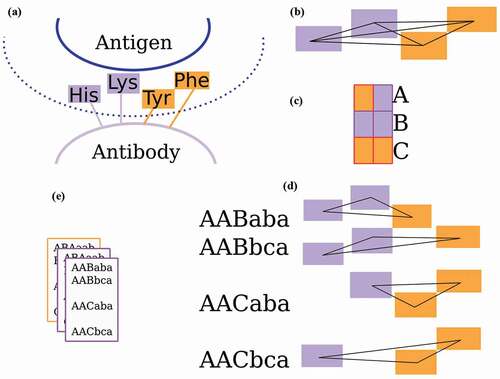
Table 4. Residue groupings for tokenization
