Figures & data
Figure 1. The cross-reactivity of MW06 to SARS-CoV-2 and SARS-CoV. (a-b) The binding ability of MW06 to SARS-CoV-2 and SARS-CoV RBD recombinant proteins was assessed by ELISA. Irrelative hIgG1 was used as a control. EC50 was labeled accordingly. (c-d) The binding kinetics of MW06 to SARS-CoV-2 and SARS-CoV RBD recombinant proteins was measured by BLI. The KD was labeled accordingly. (e-f) The ability of MW06 to block SARS-CoV-2 and SARS-CoV RBD interaction with ACE2 was evaluated by competition ELISA. Irrelative hIgG1 was used as a control. IC50 was labeled accordingly
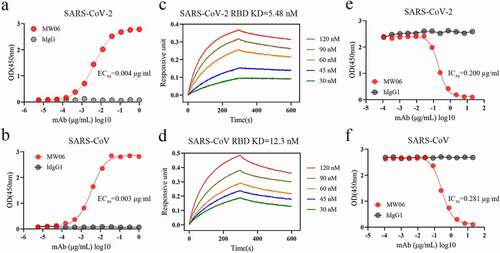
Figure 2. Neutralization activity of MW06 against SARS-CoV-2 and SARS-CoV. (a) Neutralization potency of MW06 against SARS-CoV-2 pseudovirus on Huh-7 and Vero cells, 50% neutralization titer (NT50) were labeled. (b-c) Neutralization potencies of MW06 against SARS-CoV-2 authentic virus were evaluated by CPE assay on Huh-7/hACE2 and Vero E6 cells, and plaque reduction assay on Vero E6 cells. (d) Neutralization potency of MW06 against SARS-CoV pseudovirus on Huh-7 and Vero cells. 50% neutralization titer (NT50) was labeled, accordingly
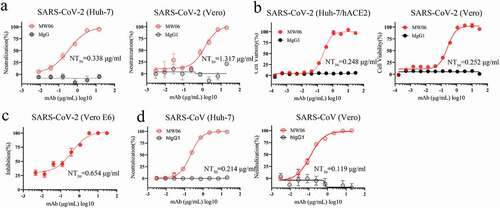
Figure 3. Determination of the complex structure of SARS-CoV-2 RBD/MW06. (a) The overall structure of MW06 Fab in complex with SARS-CoV-2 Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons, and the SARS-CoV-2 Spike RBD is shown as gray surface. (b) The epitope of MW06. The residues contacted by heavy chain, light chain, and both chains were colored in blue, light cyan, and yellow, respectively. (c) The detail of interactions between SARS-CoV-2 Spike RBD and heavy chain of MW06. (d) The detail of interactions between SARS-CoV-2 Spike RBD and light chain of MW06. The involved residues are shown as sticks with the same colors in Figure 3a. The hydrogen bonds and salt bridges are indicated as blue dashed lines. (e) Superimposition of SARS-CoV-2 RBD/MW06 with RBD/hACE2 complex (PDB code: 6LZG). The heavy chain and light chain of MW06 Fab, and the hACE2 are shown as blue, light cyan, and salmon cartoons, respectively. SARS-CoV-2 Spike RBD is represented as gray surface. (f) The MW06 and hACE2 binding surface of SARS-CoV-2 Spike RBD. The residues contacting MW06, hACE2 are colored in blue and red, respectively; the residues contacting both are colored in yellow. (g) Superimposition of SARS-CoV-2 RBD/MW06 with SARS-CoV-2 Spike trimer in “close” state (PDB code: 6VXX). One MW06 Fab is superposed to one “down” RBD (wheat) in the “close” SARS-CoV-2 Spike trimer. The binding of MW06 Fab introduces clashes (enclosed with red dashed cycle) with another RBD (light yellow). (h) Superimposition of RBD/MW06 with SARS-CoV-2 Spike trimer in “open” state (PDB code: 6VYB). The MW06 Fab is superposed to the “up” RBD (wheat). Binding of MW06 Fab introduces clashes (enclosed with red dashed cycle) with one of the “down” RBDs (light yellow). (i) Superimposition of RBD/MW06 with SARS-CoV-2 Spike trimer with 3-“up” RBDs (PDB code: 7A98). Three MW06 Fabs are superposed to three “up” RBDs in SARS-CoV-2 Spike trimer. The heavy chain and light chain of MW06 Fab are shown as blue and light cyan cartoons, respectively, and the three chains of SARS-CoV-2 Spike trimer are shown as wheat, gray, and yellow surface
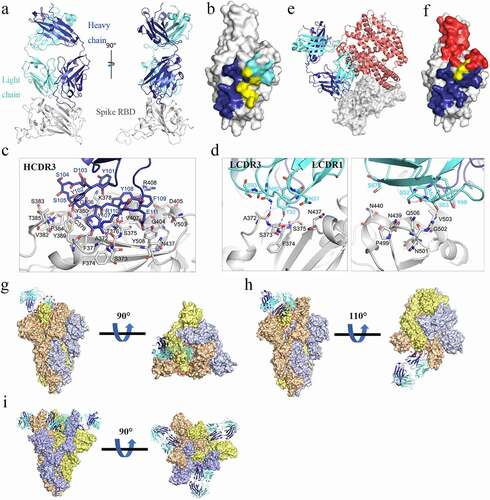
Figure 4. Conserved epitope recognized by MW06. (a) Superposition of SARS-CoV-2 Spike RBD/MW06 complex structure with SARS-CoV Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons; the SARS-CoV-2 Spike RBD is shown as gray cartoon, and the SARS-CoV Spike RBD is shown as red cartoon. (b) Sequence alignment of the Spike RBD of SARS-CoV-2 and SARS-CoV. The amino acid residues involved in interaction with MW06 Fab was indicated with pentangle. The solid pentangle indicates that the side-chains of the residue participated in the interaction while the hollow pentangle indicates main-chains. The red boxes indicated the identical amino acid residues. (c) Amino acid sequence alignment of RBDs from SARS-CoV-2, SARS-CoV and other SARS-related coronavirus strains. Epitope residues recognized by MW06 are highlighted in red. Conserved residues are indicated by small black dots on the top of the alignment
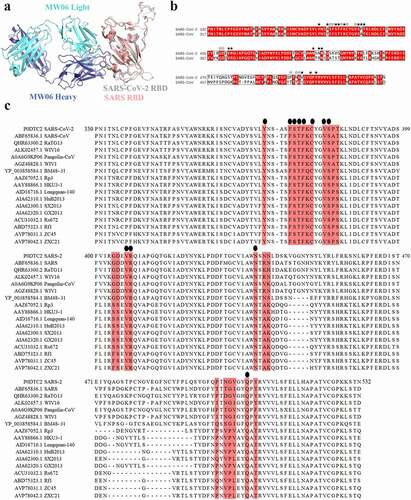
Figure 5. ADE activity of MW06. ADE of SARS-CoV-2 pseudovirus infection of nonpermissive Raji cells (a) and Daudi cells (b) by MW06 were evaluated in a luciferase assay system. SARS-CoV-2 pseudovirus were incubated with 2-fold serially diluted mAbs. The mixtures were then added into Raji cells. After 24 hours of incubation, luciferase activity was measured. MW05/IgG1 was used as positive control. (c-d) ADE of SARS-CoV pseudovirus infection of nonpermissive Raji cells and Daudi cells by MW06
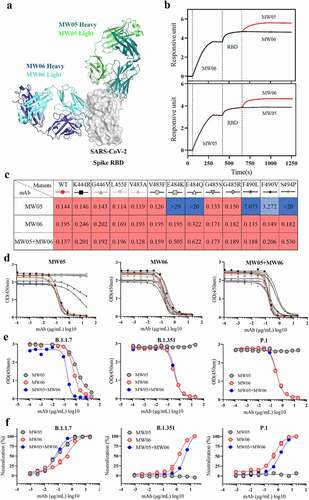
Figure 6. Cocktail of MW05 with MW06. (a) The overall structure of MW06 Fab and MW05 Fab in complex with SARS-CoV-2 Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons, the heavy chain and light chain of MW05 are shown as dark green and light green cartoons, and the SARS-CoV-2 Spike RBD is shown as gray surface. (b) Epitope competition assay was performed between MW05 and MW06. (c) Heatmap showing the ACE2/RBD blocking activity of MW05, MW06 and MW05+ MW06 were assessed by ELISA. The IC50 (μg/mL) value for each mutant is shown with red, light blue and blue indicating strong, intermediate, weak or non-detectable binding, respectively. “>20” means non-detectable activity. (d) The ACE2/RBD blocking curves of MW05 (left), MW06 (middle) and MW05+ MW06 (right). (e) The blocking ability of MW05, MW06 and the cocktail to block RBD recombination protein of SARS-CoV-2 variants strains interaction with ACE2 was evaluated by competition ELISA. (f) Neutralization potency of MW05, MW06 and the cocktail against pseudovirus of SARS-CoV-2 variants strains on Huh-7/ACE2 cells
Supplemental Material
Download Zip (467.6 KB)Data availability statement
The crystal structure for the complex of SARS-CoV-2 RBD/MW06 has been deposited in the Protein Data Bank (www.rcsb.org), under the accession no. 7DPM. All other data to support the conclusions are in the main paper or supplementary materials.
