Figures & data
Figure 1. Illustration of an antibody (a) 2-IT thiolation and payload-linker conjugation on lysine or N-terminus residues; (b) direct payload-linker conjugation on cysteine
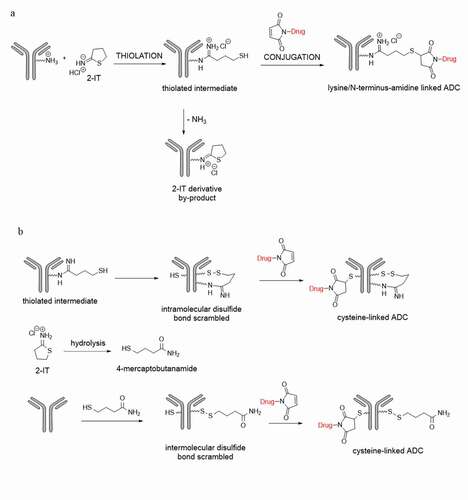
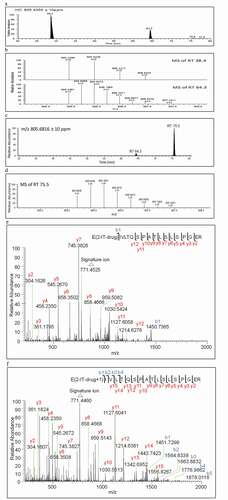
Figure 2. (a) Extracted ion chromatogram at m/z 777.2214, (b) Zoomed accurate mass spectra of the ion at m/z 777.2214 of RT 62.9, RT 64.7, RT 67.7, RT 68.6 and RT 70.1 minutes respectively, (c) Tandem mass spectrum of peak at RT 62.9, (d) Tandem mass spectrum of peak at RT 64.7, (e) Tandem mass spectrum of peak at RT 67.7, and (f) Tandem mass spectrum of peak at RT 68.5 minutes
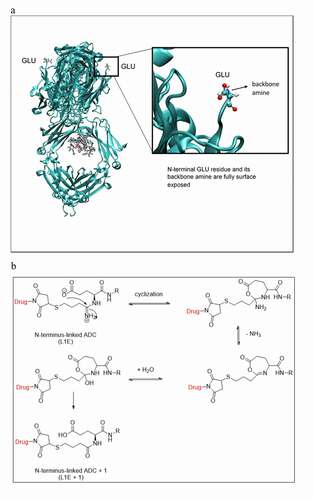
Table 1. Comparison of the solvent accessible area from modeling analysis and the relative area percent of conjugated peptides from mass spectrometry
Figure 4. Plausible formation mechanisms of the cysteine-linked conjugates. (a) (H227)K-2-IT pathways, (b) (H223)K-2-IT pathways, (c) Free thiol pathway
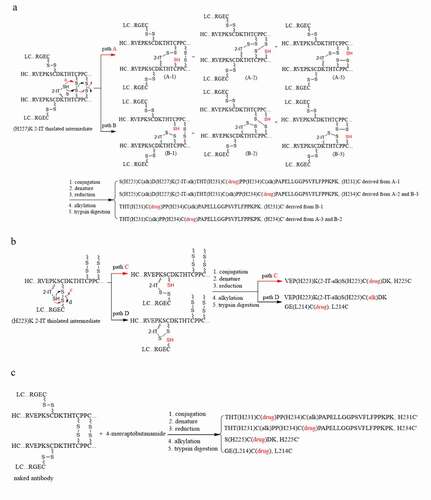
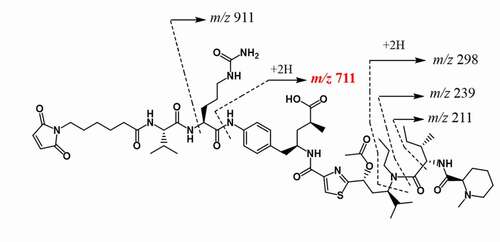
Figure 5. (a) Extracted ion chromatogram of conjugated peptide of the light chain N-terminus (L1E, XIC m/z 805.4355 ± 10 ppm), (b) Zoomed-in accurate mass spectra of the ion at m/z 805 from RT 38.4, and 64.3 minutes, respectively, (c) Extracted ion chromatogram of the hydrolyzed conjugated peptide of the light chain N-terminus (L1E + 1, XIC m/z 805.6816 ± 10 ppm), (d)Zoomed-in accurate mass spectra of the ion at m/z 805 from RT 75.6, (e) Tandem mass spectrum of N-terminal conjugated peptide (L1E) of RT 64.3 minutes, and (f) the hydrolyzed peptide (L1E + 1) of RT 75.5 minutes