Figures & data
Figure 1. Schematic outlining the bsAb discovery process and highlighting applicable stages for HTP bsAb production and screening. The main bispecific antibody discovery process phases are here defined as: 1. ‘Target ID’, identify a target pairing hypothesized and/or demonstrated to enable the desired mode of action; 2. ‘Lead Discovery’, screen and select i. mAbs against these two targets and ii. Screen and select initial Hit bsAbs exhibiting the desired mode of action; 3. ‘Lead Optimization’, engineering and more in-depth screening of initial Hit bsAbs aiming to improve functional and biophysical properties; 4. ‘early Chemistry, Manufacturing and Controls’ (early CMC), includes cell line development for large scale bsAb manufacture, formulation optimization for the final bsAb drug and more detailed assessment of bsAb product quality. Estimated format type, maximum molecule numbers, minimum purity and material quantity requirements listed in the lighter shaded box, while the primary screening purpose is stated in the darker shaded box for each panel. Unless stated otherwise, sample purity requirements are dependent upon the sensitivity of the intended screening assay to bsAb related impurities and the control samples available. At the Target Identification stage, unbiased screening of very large bsAb panels in simplified in vitro functional assays provides expanded opportunity to discover novel bsAb target pairings, which is especially powerful for obligate bsAb modes of action. For a non-obligate bsAb campaign, screening in bsAb format at Lead Discovery stage is typically not required and smaller panels are required at Lead Optimization stage as properties such as bsAb potency and selectivity correlate well with those of the parental mAbs. For obligate bsAbs, the capability to produce and screen large panels incorporating more variables at Lead Discovery stage (Option 1.2) not only increases the overall chance of successfully identifying functional hits, but also reduces the number of parameters requiring screening during Lead Optimization, potentially allowing progression directly to a Lead Panel. When larger amounts of optimization work is necessary, the capacity to screen large panels in bsAb format (Option 2.2) provides the opportunity to shorten overall timelines by exploring many interdependent factors in one panel, rather than through multiple sequential and/or parallel optimization steps (Option 2.1). At Lead Panel stage, more in depth molecule profiling is typically performed,Citation9 requiring greater amounts of ‘final format’ bsAb at higher purity, necessitating a bsAb production process incorporating increased protein expression scale and more thorough sample purification and quality control. The throughput of this process (e.g., 10 vs 100 bsAb panels) impacts how regularly projects can be accelerated directly to Lead Panel, without prior Option 2.1 or Option 2.2 screening stages. For BsAb modalities using well established bsAb targets (e.g., CD3 for T-cell engaging bsAbsCitation15,Citation19,Citation20,Citation21) potential bsAb test panel sizes will be smaller during Target ID and the availability of clinically validated mAb(s) against these targets with demonstrated potency and immunogenicity may preclude a novel mAb selections campaign during the Lead Discovery phase. Limiting the number of variable domains options on one bsAb binding module will also reduce panel sizes later on, as further optimization is then only required on the second binding module. Four panels from top to bottom highlight the main phases of a bispecific antibody discovery process. Boxes representing individual bispecific antibody screening stages are overlaid and linked into a process workflow with connecting arrows.
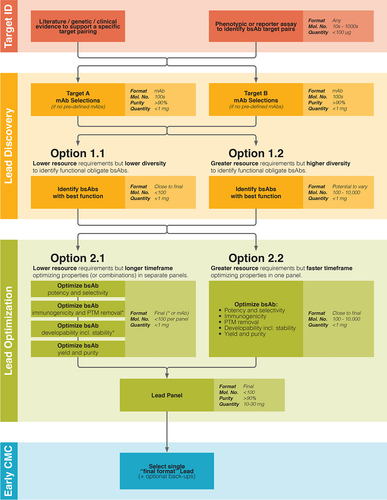
Figure 2. Overview of the major bsAb format classes and exemplar methods amenable to their HTP production. (a) Representative IgG-like (ART-Ig Citation22 and Azymetric Citation32), extended IgG-like ([2+1]-CrossMab Citation19 and IgG-(scFv)Citation2,Citation33) and linked antibody fragment formats (BiTE Citation34 and tandem dAbs Citation35) depicted. Dashed grey lines represent disulfide bonds; full grey lines, peptide linkers; blue and red rectangular modules, immunoglobulin domains within protein chains binding target 1 or target 2 respectively; yellow rectangular modules, immunoglobulin domains within a common light chain; circular module connectors, a mutation set to drive correct HC-HC pairing; triangular and square module connectors, a mutation set to drive correct HC-LC pairing. (b) Six exemplar methods amenable to HTP bsAb production: (i)Recombinant Expression, (ii) Conjugation via peptide-dAb/scFv modules,Citation36 (iii) Conjugation via split inteins,Citation37 (iv) Chemical conjugation via bis maleimide linker,Citation38 (v) Chemical conjugation via ‘click’ chemistry Citation39 and (vi) Redox recombination.Citation40 The bsAb format classes that each method can generate are marked in tickboxes. Additional features specifically depicted in (b): (i) black circles, DNA plasmids; colored plasmid overlays, DNA sequences encoding individual bsAb chains (ii) purple module, connector peptide; yellow rectangular modules, scFv with specific, high affinity to connector peptide (note this differs to key in (a)) (iii) oval purple module; N-terminal intein fragment; concave purple fragment; C-term intein fragment (iv) solid grey line; chemical or peptide linker. Top panel contains cartoons of six representative bispecific antibody formats. Bottom panel contains schematics for six different bispecific antibody high throughput production methods.
![Figure 2. Overview of the major bsAb format classes and exemplar methods amenable to their HTP production. (a) Representative IgG-like (ART-Ig Citation22 and Azymetric Citation32), extended IgG-like ([2+1]-CrossMab Citation19 and IgG-(scFv)Citation2,Citation33) and linked antibody fragment formats (BiTE Citation34 and tandem dAbs Citation35) depicted. Dashed grey lines represent disulfide bonds; full grey lines, peptide linkers; blue and red rectangular modules, immunoglobulin domains within protein chains binding target 1 or target 2 respectively; yellow rectangular modules, immunoglobulin domains within a common light chain; circular module connectors, a mutation set to drive correct HC-HC pairing; triangular and square module connectors, a mutation set to drive correct HC-LC pairing. (b) Six exemplar methods amenable to HTP bsAb production: (i)Recombinant Expression, (ii) Conjugation via peptide-dAb/scFv modules,Citation36 (iii) Conjugation via split inteins,Citation37 (iv) Chemical conjugation via bis maleimide linker,Citation38 (v) Chemical conjugation via ‘click’ chemistry Citation39 and (vi) Redox recombination.Citation40 The bsAb format classes that each method can generate are marked in tickboxes. Additional features specifically depicted in (b): (i) black circles, DNA plasmids; colored plasmid overlays, DNA sequences encoding individual bsAb chains (ii) purple module, connector peptide; yellow rectangular modules, scFv with specific, high affinity to connector peptide (note this differs to key in (a)) (iii) oval purple module; N-terminal intein fragment; concave purple fragment; C-term intein fragment (iv) solid grey line; chemical or peptide linker. Top panel contains cartoons of six representative bispecific antibody formats. Bottom panel contains schematics for six different bispecific antibody high throughput production methods.](/cms/asset/9c0eb149-91e1-457b-a616-29038e10dd4f/kmab_a_2311992_f0002_oc.jpg)
Table 1. List of liquid chromatography methods employed to characterize and/or purify IgG-like bispecific antibodies. Examples of bispecific antibody formats analyzed (or purified) using these standalone methods are provided under ‘offline MS’, while examples where these methods have been used in tandem with mass spectrometry are listed under ‘online MS’.
