Figures & data
Figure 1. Sequence analysis of Arabidopsis SP1, SPL1 and SPL2 and human MAPL proteins. (A) Amino acid sequence alignment of SP1, SPL1, SPL2 and MAPL performed by the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Identical and similar residues are shaded. Predicted transmembrane (TMD) and RING domains are indicated by boxes. (B) Phylogenetic analysis of plant SP1-related proteins and human MAPL by Phylogeny.fr (http://www.phylogeny.fr/). At, Arabidopsis thaliana. Os, Oryza sativa. Hs, Homo sapiens. Scale bar, 0.6 amino acid substitutions per site. Branch support values are shown as percentage.
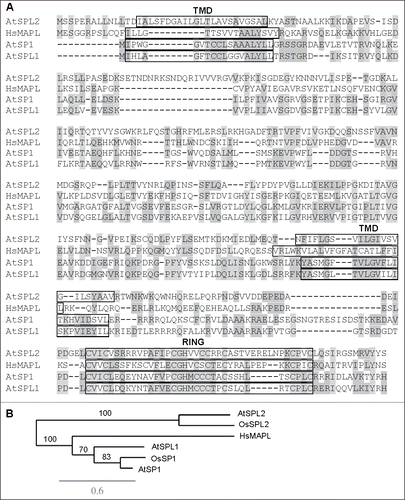
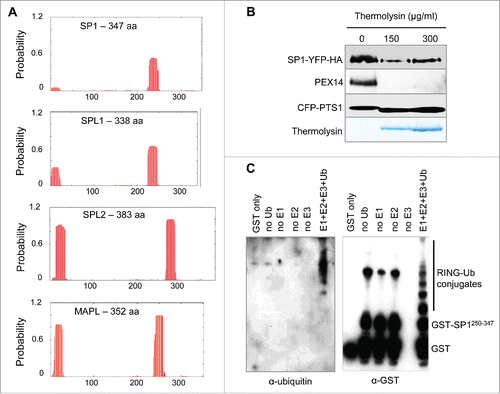
Figure 2. Characterization of the SP1 protein. (A) Transmembrane domain (TMD) analysis of Arabidopsis SP1, SPL1, SPL2 and human MAPL by the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/). Columns indicate potential TMDs. Y axis represents the probability of TMDs, and x-axis indicates the length of the analyzed proteins. (B) Protease protection assay to determine membrane topology of SP1 on the peroxisome. Peroxisomes were isolated from Arabidopsis plants co-expressing SP1-YFP and CFP-PTS1 (peroxisome targeting signal type 1; SKL), which had been generated in our previous study,Citation18 treated with thermolysin, and subjected to immunoblot analysis with α-GFP and α-PEX14 antibodies, using protocols that we used previously.Citation29 Here, 200 μl purified peroxisomes was treated respectively with 0, 150 or 300 μg/ml of thermolysin in an incubation buffer containing 50 mM Hepes/NaOH, pH 7.5, 0.33 M sorbitol, and 0.5 mM CaCl2. Reactions were performed at 4°C for 30 min, and stopped by 5-min incubation on ice with 5 mM EDTA, followed by immunoblot analysis. The thermolysin bands were Coommassie Blue-stained. (C) In vitro ubiquitination assays using a previously published protocol.Citation25 Immunoblot analyses were performed using anti-Ubiquitin (1:10,000; Invitrogen) and anti-GST (1:150; Sigma-Aldrich) antibodies.