Figures & data
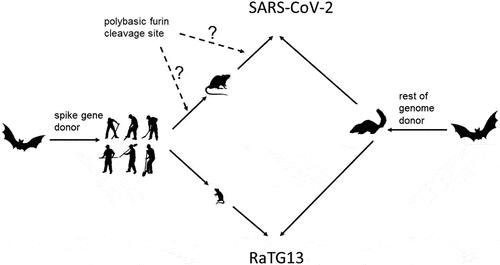
Figure 1. Distances between SARS-CoV-2 pentapeptide vocabulary and pentapeptide vocabularies of the main clades of mammals.
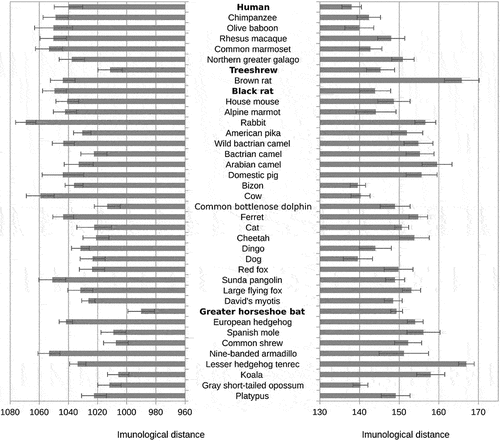
Figure 2. Distances between SARS-CoV-2 hexapeptide vocabulary and hexapeptide vocabularies of the main clades of mammals.
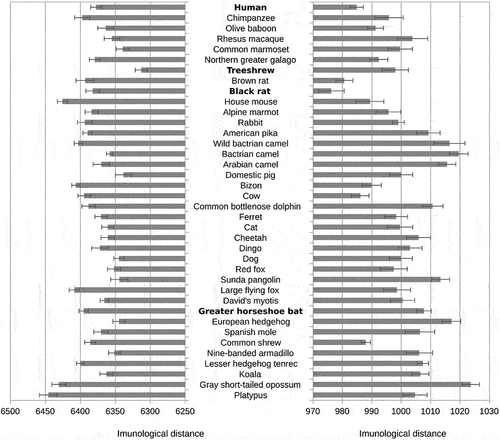
Figure 3. Probable origins of SARS-CoV-2 and RaTG13.