Figures & data
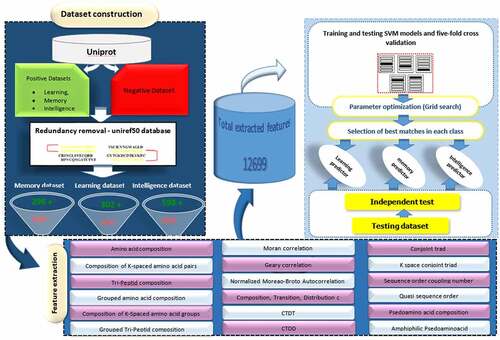
Table 1. Summary of 12,669 extracted protein features that were used in SVM-based model development. More details are provided in Supplementary File S2.
Table 2. Accuracy metrics of train and test datasets for intelligence model. The optimum γ parameter value of kernel function of SVM was chosen using a grid-search technique based on five-fold cross-validation.
Table 3. Accuracy metrics of train and test datasets for memory model. The optimum γ parameter value of kernel function of SVM was chosen using a grid-search technique based on five-fold cross-validation.
Table 4. Accuracy metrics of train and test datasets for learning model. The optimum γ parameter value of kernel function of SVM was chosen using a grid-search technique based on five-fold cross-validation.
Table 5. Summary results of evaluation of the 10,000 new negative protein sequences by Intell_Pred.
Table 6. Summary results of evaluation of the new annotated protein sequences related to intelligence in Uniprot database by Intell_Pred (169 protein sequences were evaluated including 85 and 84 proteins belong to memory and learning classes, respectively) .
Table 7. Summary results of evaluation of the plant candidate proteins using Intell_Pred.