Figures & data
Figure 1. The work flow of spectral-spatial classification with EMAP. The dotted lines indicate the possibility of switching between supervised and unsupervised feature reduction. An optional feature reduction step can be used to reduce the dimensionality of EMAP before classification.
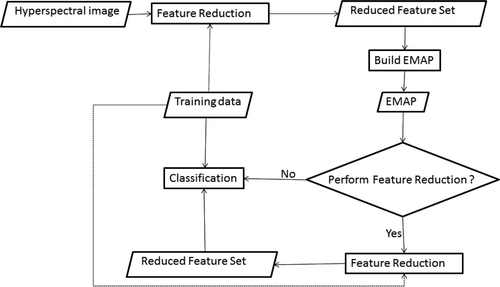
Figure 2. Indian Pine dataset. (a) False colour representation of the Indian Pines image, (b) test and (c) training sets.
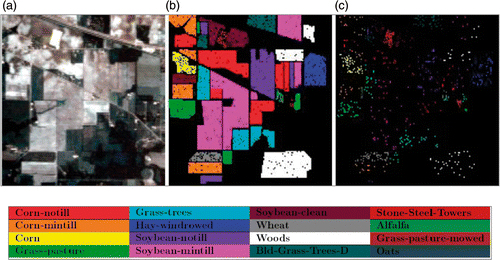
Table 1. Number of training and test pixels used for the Indian Pines image.
Figure 3. Pavia University dataset. (a) False-colour representation of the University of Pavia image, (b) test and (c) training set.
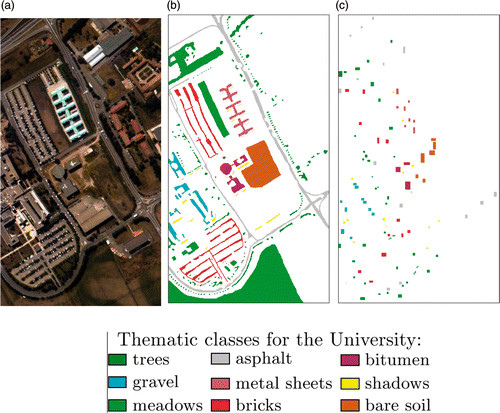
Table 2. Number of training and test pixels used for the Pavia image.
Table 3. Accuracies (%) for classification of Indian Pines image. The best results in terms of accuracy are marked in bold.
Table 4. Accuracies in percentage for classification of the University of Pavia image. The best results in terms of accuracy are marked in bold.
Figure 4. Classification maps for Indian Pines data with RF classifier using (a) Original data (b) PCA, (c) KPCA, (d) DAFE, (e) DBFE and (f) NWFE.
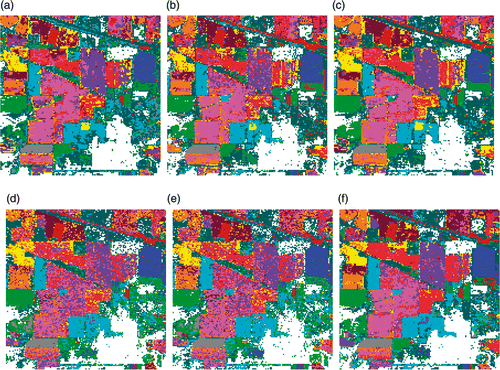
Figure 5. Classification maps for Indian Pines data with SVM classifier using (a) Original data (b) PCA, (c) KPCA, (d) DAFE, (e) DBFE and (f) NWFE.
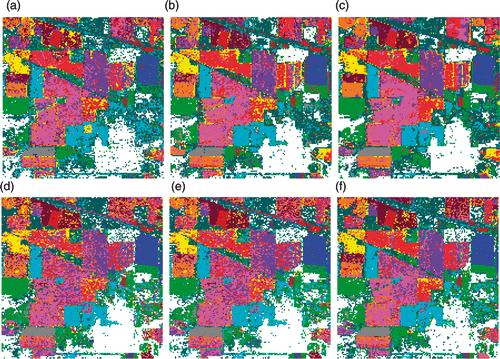
Figure 6. Classification maps for Indian Pines data with RF classifier using EMAPs of (a) PCA, (b) KPCA, (c) DAFE, (d) DBFE and (e) NWFE, and feature reduction applied on EMAP using (f) NW-NW and (g) KP-NW.
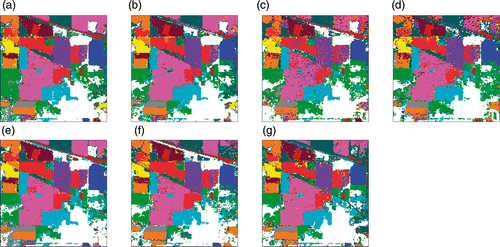
Figure 7. Classification maps of Indian Pines data with SVM classifier using EMAPs of (a) PCA, (b) KPCA, (c) DAFE, (d) DBFE and (e) NWFE, and feature reduction applied on EMAP using (f) NW-NW and (g) KP-NW.
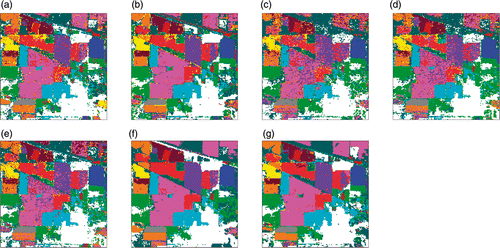
Figure 8. The ground reference data for the Indian Pines image. The segments shown in white boxes are found to be difficult to classify.
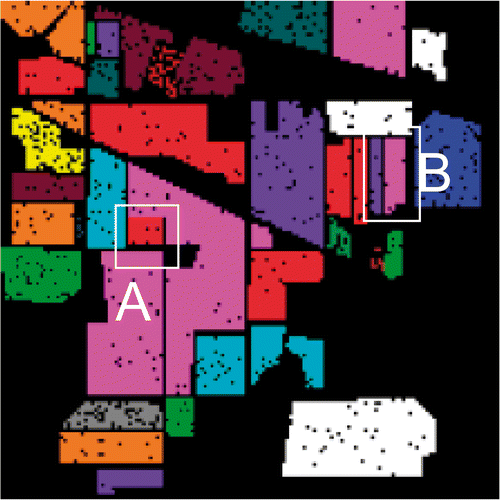
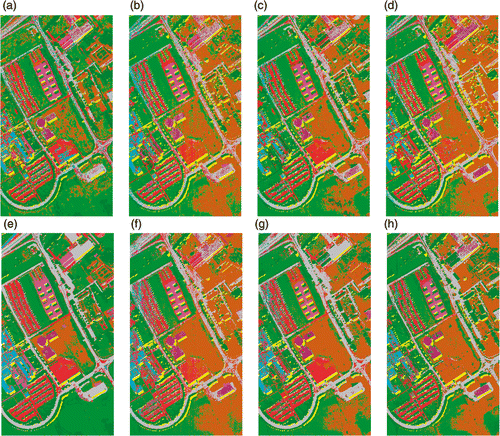
Figure 9. Classification maps for University of Pavia data with RF classifier using (a) Original data (b) PCA, (c) KPCA, (d) DAFE, (e) DBFE and (f) NWFE.

Figure 10. Classification maps for University of Pavia data with SVM classifier using (a) Original data (b) PCA, (c) KPCA, (d) DAFE, (e) DBFE and (f) NWFE.

Figure 11. Classification results of University of Pavia data with RF classifier using EMAPS of (a) KPCA, (b) DBFE, (c) NWFE and feature reduction applied on EMAP using (d) DB-DB.

Figure 12. Classification maps of University of Pavia data with SVM classifier using EMAPS of (a) KPCA, (b) DBFE, (c) NWFE and feature reduction applied on EMAP using (d) DB-DB.

Table 5. Accuracies in percentage for classification of Pavia image with 30 training pixels per class. The best results in terms of accuracy are marked in bold.
Table 6. Accuracies in percentage for classification of Pavia image with 100 training pixels per class. The best results in terms of accuracy are marked in bold.
Figure 13. A snapshot of University of Pavia region from Google Maps (accessed 02 November 2011). A big part of the image remains unchanged from the date of acquisition of the ROSIS image. The boxes delineates specific areas considered in the discussion to visually compare the geometric stability of the classification results. Imagery ©2011 DigitalGlobe, GeoEye.
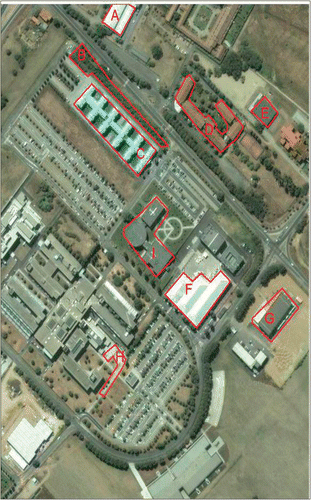
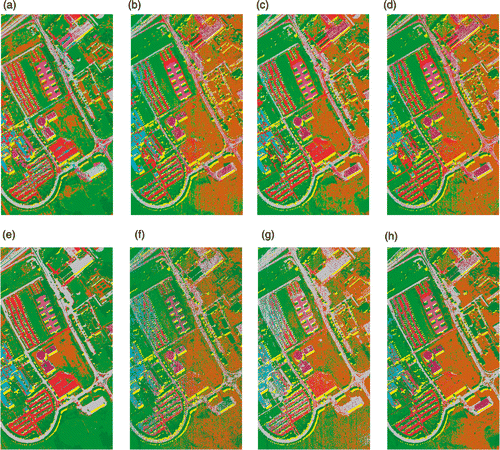
Figure 14. Classification maps of University of Pavia data using 30 training samples per class with RF classifier using (a) KPCA, (b) DAFE, (c) DBFE, (d) NWFE, (e) KPCA p , (f) DAFE p , (g) DBFE p and (h) NWFE p .
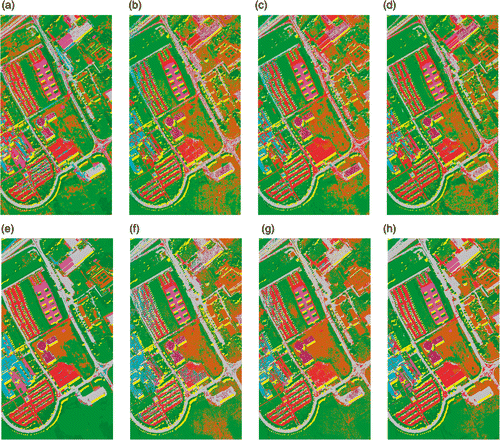
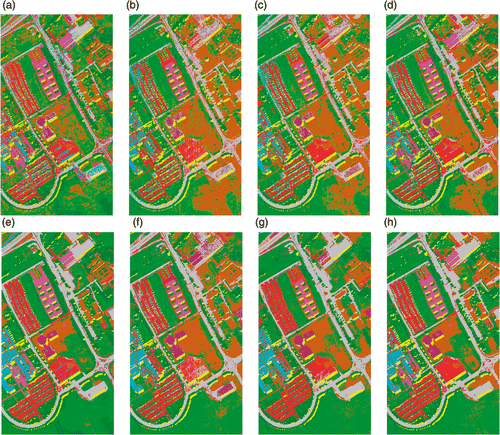
Figure 15. Classification maps of University of Pavia data using 30 training samples per class with SVM classifier (a) KPCA, (b) DAFE, (c) DBFE (d) NWFE, (e) KPCA p , (f) DAFE p , (g) DBFE p and (h) NWFE p .