Figures & data
Figure 1. (A) The luminal microbiome (represented by fecal samples) has higher microbial diversity than the mucosal microbiome (represented by biopsy samples) as determined by paired t-test. ***P < 0.0005. (B) PCoA plot of unweighted UniFrac distance, showing that the bacterial communities of the stool samples (blue dots) were distinct from those in the biopsy samples (red dots). (C) Relative abundance of taxonomic groups averaging across stool and biopsy samples, with the 10 most abundant phyla being represented. (D) Average UniFrac distance calculated between the luminal and mucosal microbial communities was greater than the average UniFrac distance calculated for different microbial communities between different individuals. (E) Bacterial taxa identified to be differentially abundant in the mucosal and luminal microbiome by LEfSe, at a logarithmic LDA threshold score of 2.0.
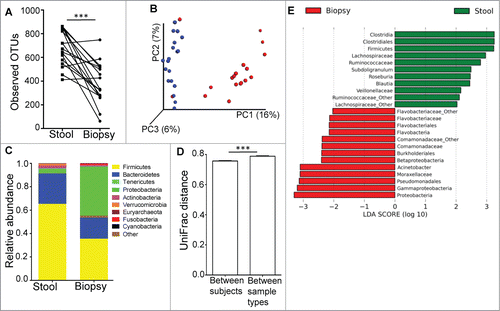
Figure 2. (A) Bacterial taxa that were significantly enriched in the mucosa of participants undergoing routine screening colonoscopy and were previously identified to be differentially enriched in mucosa of patients with colitis. (B) Bacterial taxa that were significantly enriched in the luminal microbiome of participants undergoing routine screening colonoscopy and were previously identified to be reduced in the mucosa of inflamed tissue of UC patients relative to uninflamed normal mucosa. Paired t-tests: *P < 0.05, ***P < 0.0005.
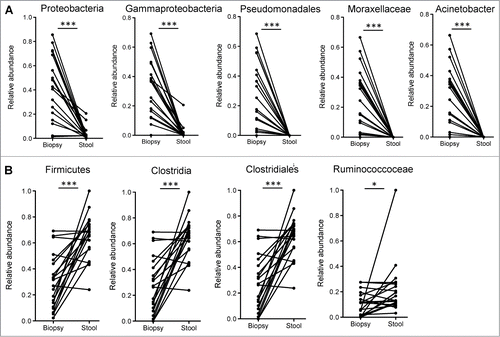
Figure 3. (A) Supervised analysis with LEfSe (Log LDA > 2.00) showed functional pathways that were differentially enriched in the mucosal microbiome and the luminal microbiome. (B) Paired analyses of stool and biopsy samples from the same individuals found lipid and amino acid metabolism pathways to be enriched in the mucosal microbiome, while carbohydrate and nucleotide metabolism pathways, as well as genetic information processing pathways to be more abundant on the luminal microbiome. Paired t-tests: **P < 0.005, ***P < 0.0005.
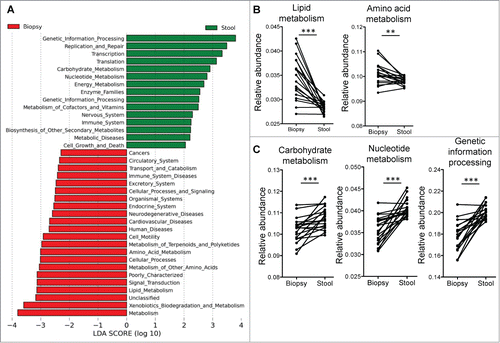
Figure 4. Spearman correlation analysis of the differences in relative abundance values of bacterial taxa from both data sets. The values along the B-S axis represented the differences in relative abundance of bacteria between biopsy and stool samples, while the values along the I-N axis represented the differences in relative abundance of bacteria between inflamed and normal biopsy tissues. Each data point is representative of a single bacteria taxa. The correlation analysis was performed at both the phylum (A) and genus (B) levels, with the data points for the phyla Proteobacteria and Firmicutes, as well as the genus Acinetobacter being highlighted. The same analysis was repeated with relative abundance values of predicted metabolic pathways (C). (D) The enrichment of Acinetobacter genus in mucosal biopsy was validated by performing qPCR on biopsy and stool samples. Mann-Whitney test: ***P < 0.0005.
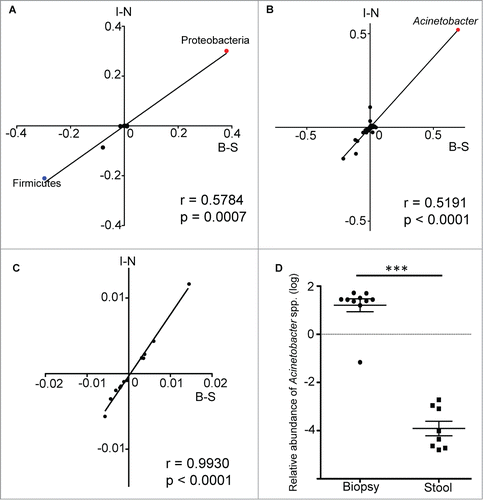
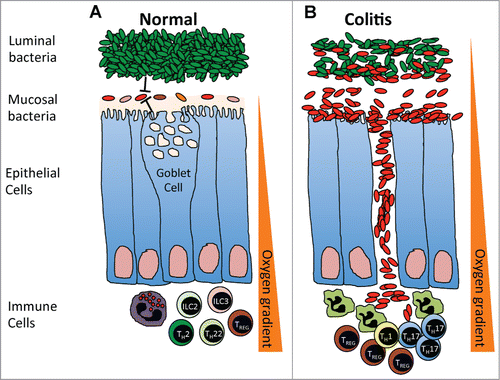
Figure 5. (A) In a steady-state, the mucosal microbial communities are kept in check by the luminal bacteria, intestinal immune response and barrier function. An oxygen gradient exists within the intestinal environment, with the mucosal interface being largely aerobic and the intestinal lumen largely anaerobic. (B) During colitis, the intestinal barrier breaks down, increasing the oxygen content within the intestinal lumen and leading to the expansion of aerotolerant mucosal microbial communities, which spill over into the intestinal lumen and translocate across the intestinal epithelial barrier surface to trigger a strong anti-bacterial response.