Figures & data
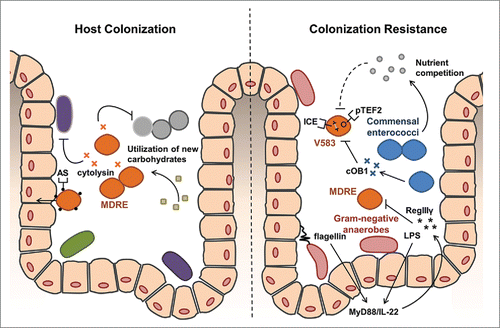
Figure 1. The convergence of acquired traits that can facilitate host colonization by VRE and mechanisms limiting it. Left panel. Several traits have evolved in drug-resistant enterococcal lineages that may work cooperatively to enhance the gastrointestinal colonization and persistence. Cytolysin, a bipartite bacteriocin, has broad spectrum activity against Gram-positive bacteria that likely eliminates commensals in the local environment to reduce competition.Citation35 The surface protein aggregation substance (AS) can promote adherence and invasion of intestinal epithelial cells which promote intestinal persistence.Citation1 Additional carbohydrate uptake and metabolism functions associated with the enterococcal pathogenicity island may allow for more efficient exploitation of available carbohydrate resulting from depletion of normal gut flora.Citation36 Right Panel. Gram-negative anaerobic commensals provide generalized resistance against VRE through the surface factors lipopolysaccharide (LPS) and flagellin activate MyD88 and IL-22 signaling pathways to stimulate host production of the defensin RegIIIγ, which has broad activity against Gram-positives.Citation37 Commensal enterococci may also exert more specific resistance against VRE colonization by competing for shared nutrients.Citation21 In some cases, the accumulation of mobile genetic elements, like the conjugative plasmid pTEF2 and an integrated chromosomal element (ICE), can make VRE incompatible with commensal strains that produce otherwise harmless peptide pheromones.Citation21