Figures & data
Figure 1. Schematic for antibiotic and probiotic administration. Sixteen C57BL/6 mice received antibiotic treated water for 2 weeks in duration. Mice were then split evenly into 4 treatments: 1) continued antibiotics, 2) continued antibiotics with gavaged probiotics 3) regular water with gavaged probiotics, 4) regular water. Control animals were maintained on regular water throughout the experiment. Mice were euthanized and contents of cecum and large intestine collected.
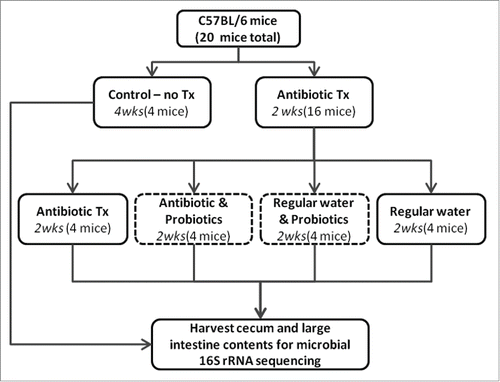
Figure 2. Comparison of cecal/intestinal microbial community structure by rank ordering relative abundance of bacterial families following antibiotic and probiotic treatment. More than 2 million OTU tags were retrieved from sequenced DNA and relative abundance at the taxonomic family level was calculated. Family level abundance was pooled for each experimental group. Any bacterial family above 1% total in any single animal lead to inclusion in figure for all groups. Control mice did not receive antibiotics. Ab mice were on antibiotics for all 4 weeks. Ab/Pb mice were on antibiotics for 4 weeks and the last 2 weeks were supplemented with gavaged probiotics. Recovery mice were switched to regular water with or without probiotic treatment.
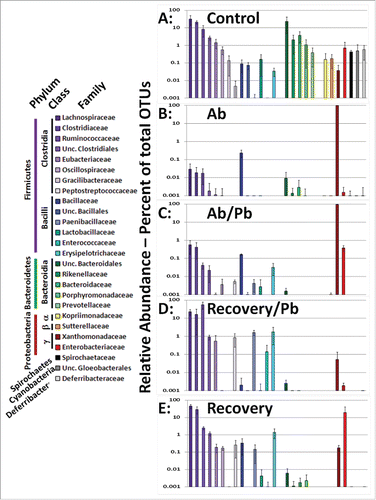
Figure 3. Impact of antibiotic treatment and probiotic supplementation on microbial diversity in the mouse intestine. Diversity analysis utilized the QIIME pipeline. Alpha diversity was determined based on rarefied analysis of A) Chao1, B) Phylogenetic diversity, C) Shannon index, and D) Observed species abundance. Standard deviations among group members are provided. Intergroup comparisons were made based on principal coordinate analysis of unweighted UniFrac distances (E). In the biplot, the 5 most prevalent taxa driving the differences in the PCoA are provided (F = family level, O = order level). Taxon specific spheres vary in size based on mean relative abundance in all groups. (** = p < 0.01)
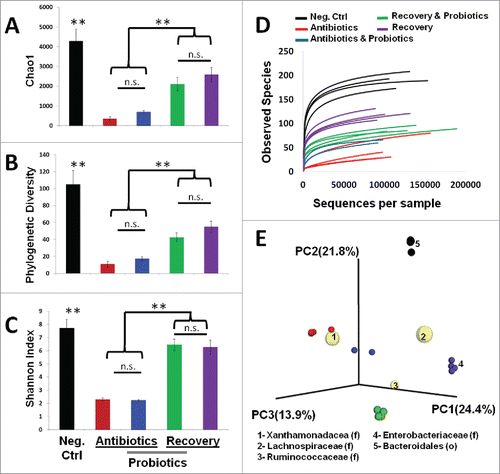
Figure 4. Genus level diversity of dominant 15 taxa among treatment groups. The top 5 genera from each treatment group were consolidated into a pool of 15 dominant genera. These dominant genera make up the majority of OTUs from each mouse, average OTU coverage ± standard deviation displayed above pie charts. A) Control animals B) Antibiotics for 4 weeks C) Antibiotics for 4 weeks supplemented with probiotics D) Recovery from antibiotics supplemented with probiotics E) Recovery from antibiotics on regular water.
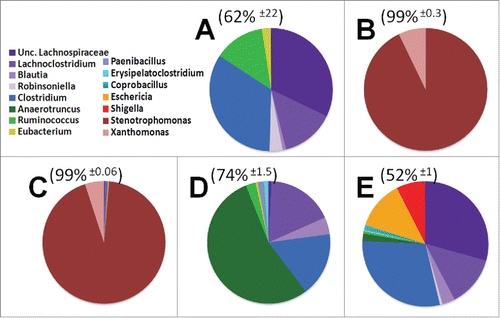
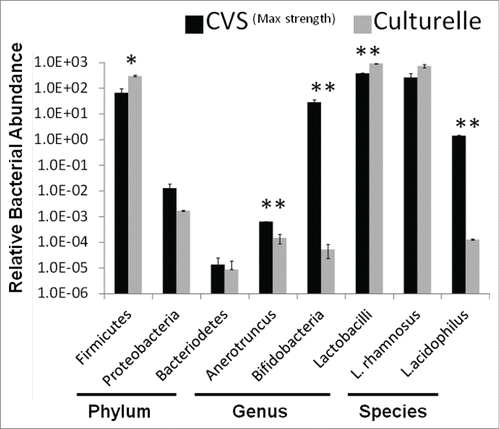
Figure 5. Relative abundance of bacterial taxa in probiotics. Quantitative PCR was used to determine the relative abundance of bacteria in probiotic supplements to determine ‘purity’. Probiotic mixture CVSMax Strength was compared to mono-culture probiotic Culturelle™. Probiotics were screened at the phylum, genus, and species level using taxa specific PCR primers. (* = p < 0.05, ** = p < 0.01) Representative of 3 independent experiments.