Figures & data
Figure 1. Modulation of bacterial genera by cefprozil. Average change in abundance ranking between day 7 and day 0 samples for exposed participant. Only the 13 genera for which relative abundance was statistically significant between day 7 and day 0 after antibiotics treatment are displayed (p < 0.05 after Benjamini & Yekutieli false discovery rate correction, see Raymond et al.Citation7 for details). Error bars represent confidence interval at 95%.
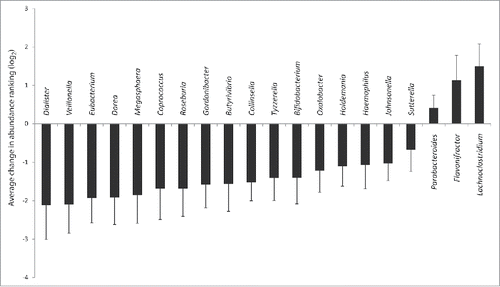
Figure 2. Comparison of complete assembled metagenomes between A) day 7 vs day 0 and B) day 90 vs day 0 for exposed participants and controls. Horizontal lines indicate the mean of groups. Participants enriched in E. cloacae at day 7 are marked with stars (*).
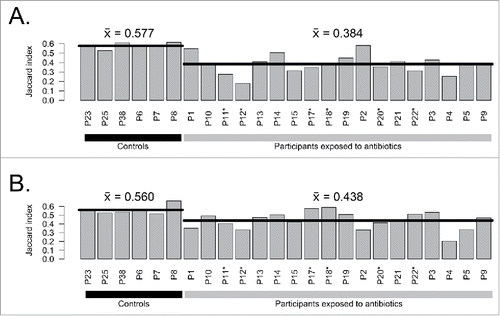
Figure 3. Comparison of total k-mers content between A) day 7 and day 0 and B) day 90 and day 0. Counts are normalized by the number of reads sequenced per sample. Participants enriched in E. cloacae at day 7 are marked with stars (*).
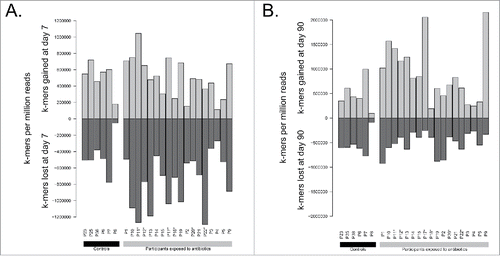
Figure 4. Comparison of resistance gene related k-mers between A) day 7 vs day 0 and B) day 90 vs day 0 for exposed participants and controls. Horizontal lines indicate the mean of groups. Participants enriched in E. cloacae at day 7 are marked with stars (*).
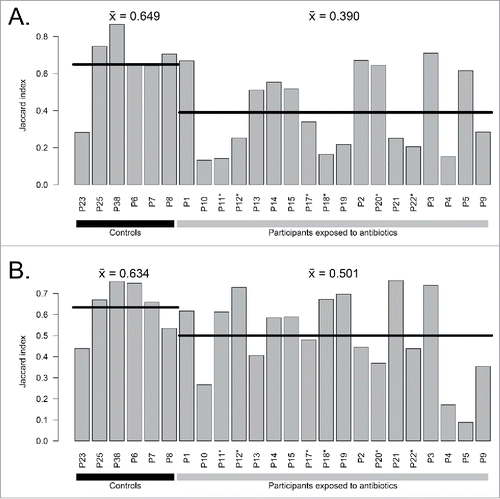
Figure 5. Comparison of k-mers related to antibiotic resistance between A) day 7 and day 0 and B) day 90 and day 0. Counts are normalized by the number of reads sequenced per sample. Participants enriched in E. cloacae at day 7 are marked with stars (*).
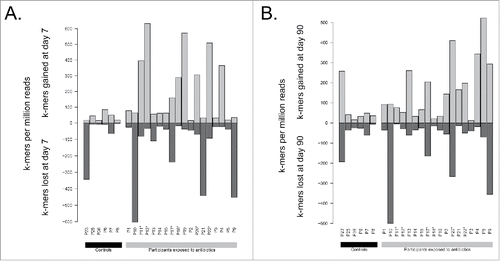
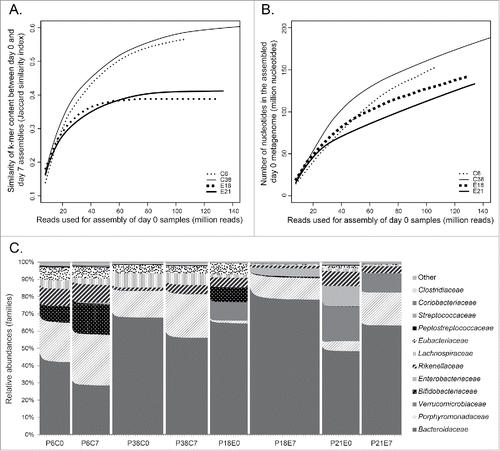
Figure 6. Influence of sequencing depth on comparison, assembly and profiling of metagenomes for 4 participants (controls 6 and 38, and exposed to antibiotic 18 and 21). A) Comparison of k-mer content of day 0 assemblies with day 7 assemblies using Ray Surveyor. Day 7 assemblies were performed using all the reads sequenced while day 0 assemblies were performed by subsampling reads using 8 million reads increments. Similarity was calculated using the Jaccard similarity index. B) Influence of the number of reads used for assembly on the number of nucleotides assembled into contigs for day 0 samples. Subsampling of reads was in 8 million reads increments. C) Taxonomical profiling at the family rank using Ray Meta 2.3.1 on subsampled reads. Subsampling increments of 8 million nucleotides are indicated by dashes under the plot.