Figures & data
Table 1. Transporters and junctional proteins expressed in gut endothelial cells. Gene list of transporters and adherens and tight junction proteins with an expression value greater than 140 (normalized read count) in CD45−CD31+CD105+LYVE1− gut blood endothelial cell.
Figure 1. The Gut Vascular Unit. Confocal images of C57BL/6 mice intestine stained with CD34 (green) to identify blood endothelium and GFAP (red), marker of enteric glial cells (left panels; scale bars: 20 and 10 μm) or desmin (red) that marks perycites (scale bar 50 μm). In each section, nuclei were stained with DAPI (blue). Both cell types are in close proximity to CD34-positive endothelial cells, that together for the gut-vascular unit (GVU).
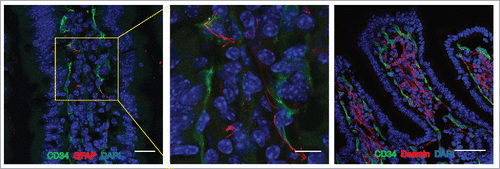
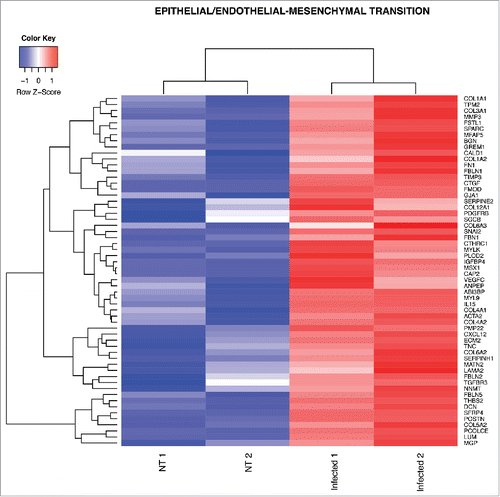
Figure 2. Heat map from hierarchical clustering representing DEGs belonging to SHH pathway. CD45−CD31+CD105+LYVE1− blood endothelial cell were purified by FACS sorting from small intestine of mice orally infected with S. typhimurium for 6 hours. Differential expression analysis was performed to investigate DEGs between infected and untreated mice. The analysis was carried out using DESeq2 R package. After read counts normalization across the samples, the expression of each gene was tested between the two conditions and to avoid false positive expression due to technical sequencing errors only high-quality transcript counts have been analyzed (filter transcript with 0 read count). DEGs with corrected p-value (FDR) less than 0.05 were considered as statistically different between the two conditions. The heat map shows DEGs that are annotated as SHH pathway genes according to Gene Ontology from MGI (GO:0007224). Red upregulated DEGs and blue down-regulated DEGs. NT=untreated.
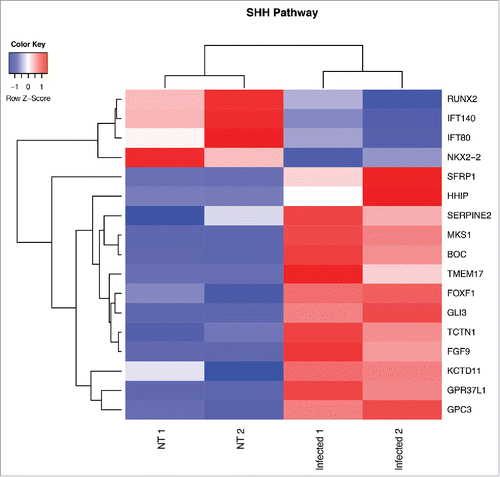
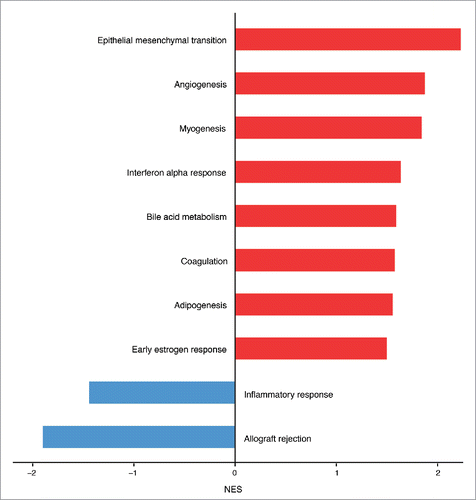
Figure 3. Hallmark molecular functions enriched in endothelial cells upon Salmonella infection. The bars plot shows the statistically significant GSEA Hallmark gene sets (FDR<0.05) that were found enriched in correlation (red bars) or inverse correlation (blue bars) with infected phenotype. The genes were ranked using the Signal2Noise metric (available in GSEA) by taking as reference the treated samples. Gene sets with a FDR lower than 0.05 have been considered statistically significant. The bars were ordered by normalized enrichment score (NES) that indicates the strength of the enrichment.