Figures & data
Table 1. Participant Demographics and Post-surgical Clinical Data.
Figure 1. Structure of the intestine. (A) Loop ileostomy and (B) following reanastomosis. Block arrows denote presence and direction of luminal contents flow. Tissue located above dashed lines represent areas of intestine removed before reanastomosis and form the specimen acquired for our research.
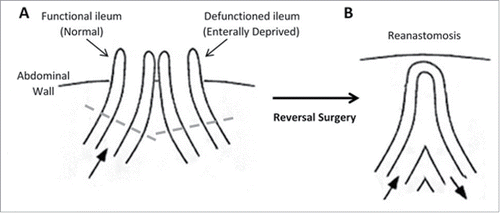
Figure 2. Histological analyses of function and defunctioned ileum. (A) Representative H&E stained sections, magnification 4x. (B) Average villous height ± SEM (n = 9, p = 0.0004) (C) Paired villous height.
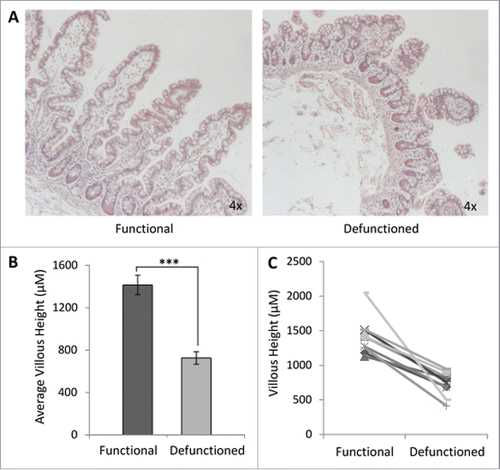
Figure 3. Bacterial enumeration of (A) average- and (B) absolute-16S rRNA gene copy numbers per gram of mucosal tissue in functional and defunctioned intestine (n = 27; p = 0.0003).
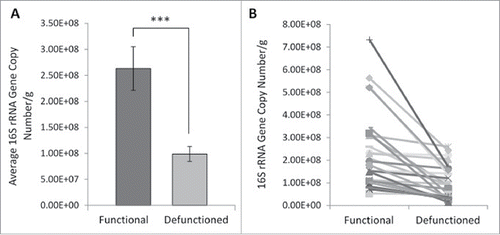
Figure 4. (A) Example of denaturation gradient gel electrophoresis (DGGE) profiles. Band classes are depicted using characters a through h and corresponding bands are enclosed. F, functional ileum; D, defunctioned ileum. (B) Microbial diversity in functional and defunctioned ileum expressed as total number of bands in DGGE profiles (n = 11; P = 0.04). (C) Purified and sequenced band classes expressed as a percentage presence in DGGE profiles across all patients. Characters a-h correspond to the band classes highlighted in A. Inclined, corresponding assigned bacterial genus per band class.
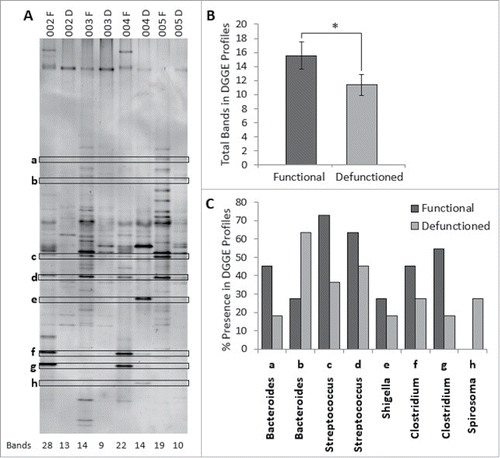
Figure 5. Hierarchical cluster analysis of DGGE profiles represented in graphical form as an UPGMA dendogram. F: functional ileum, D: defunctioned ileum (n = 11).
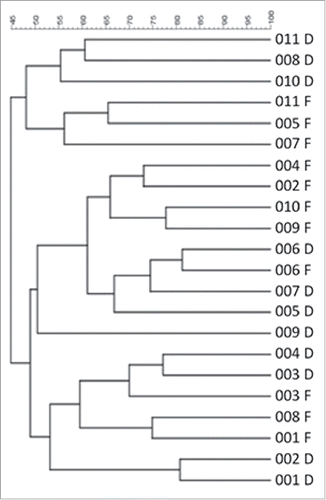
Figure 6. Percentage change of phyla abundance in defunctioned ileum relative to functional. Data normalized to universal 16S rDNA primers (Firmicutes (n = 18, p = 0.02), Bacteroiodetes (n = 18, NS), y-Proteobacteria (n = 9, p ≤ 0.05).
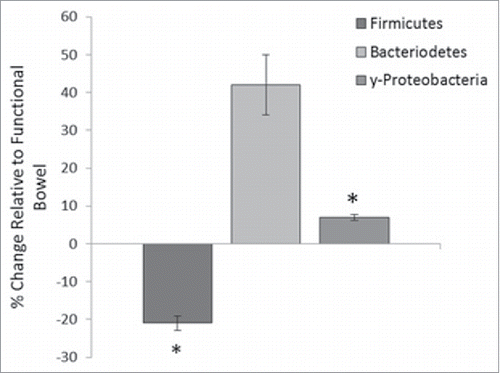
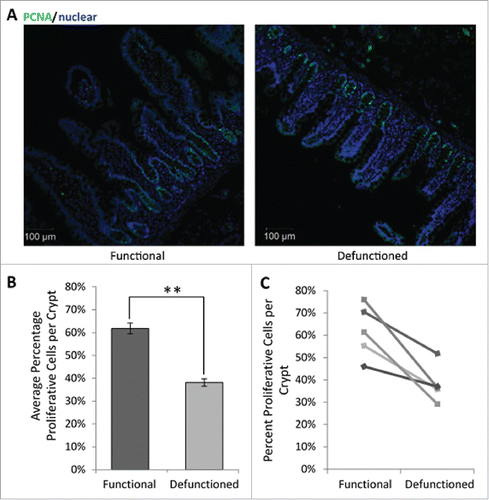
Figure 7. (A) Representative immunofluorescent PCNA labeling (green) to measure proliferation. All nucleated cells, counterstained using Hoechst 33342, are colored blue. Magnification 10x. (B) Average percent proliferating PCNA positive cells/crypt ± SEM (n = 5; p = 0.01). (C) Paired percent PCNA-positive cells per crypt in the functional and defunctioned intestine.