Figures & data
Figure 1. (A) General mechanism of glycyl radical enzymes (GREs). A partner radical S-adenosylmethionine (SAM) activating enzyme (AE) first abstracts a hydrogen atom from the α-carbon of a conserved active site glycine residue on the GRE to generate a carbon-centered radical. The glycine-centered radical is proposed to abstract a hydrogen atom from an essential active site cysteine residue to generate a thiyl radical intermediate that initiates reaction with the substrate. Further reaction of the substrate-based radical generates a product-based radical, which then abstracts a hydrogen atom from the essential cysteine residue to regenerate the thiyl radical. (B) Previously characterized activities of 4-hydroxyproline dehydratase (HypD) and Δ1-pyrroline-5-carboxylate reductase (P5CR) from Clostridioides difficile 70-100-2010.
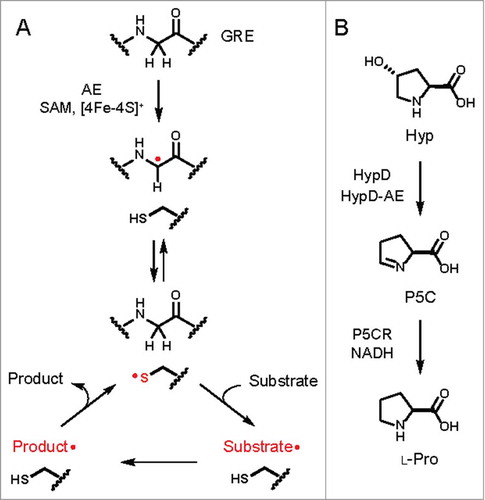
Figure 2. Characterized pathways for Hyp metabolism. (A) Pathway for Stickland fermentation using l-proline (Pro) as an electron acceptor. The product of Pro reduction, 5-aminovalerate, can be further oxidized and reduced by certain Clostridiales to generate short-chain fatty acids and ammonia. (B) HypD provides an anaerobic route for Hyp metabolism. (C) In mammals, Hyp is oxidized to glyoxylate and pyruvate in a multistep pathway. (D) An oxidative pathway for Hyp metabolism generates α-ketoglutarate (α-KG) and ammonia as sources of carbon and nitrogen.
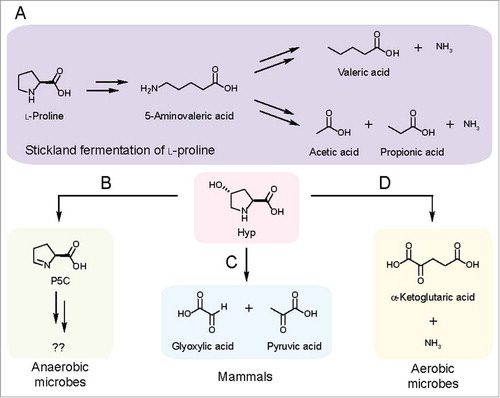
Figure 3. Growth curves and metabolite analysis of two HypD-encoding Clostridiales cultures reveal an increase in downstream Stickland fermentation metabolites accompanying Hyp consumption. (A, D) Enhanced growth was observed for both C. difficile 630Δerm and T. glycolicus DSM 1288 upon supplementation of a previously reported medium with either Pro or Hyp (20 mM final concentration).Citation5 Downstream metabolites were quantified at various timepoints in cultures supplemented with Pro (B, E) or Hyp (C, F). Experiments were set up as previously describedCitation5 with the following modifications: 200 µL cultures were grown in 96-well plates and OD600 measurements were taken using a PowerWave HT Microplate Spectrophotometer (BioTek). Samples were collected at various time points for metabolite quantification by liquid chromatography tandem-mass spectrometry.Citation5 5-Aminovalerate was measured using the Hyp detection method by monitoring precursor and product ions of m/z 118.1 → m/z 101.1. Sample peak areas are normalized to the corresponding standard. Data points are shown as mean ± standard deviation with n = 5.
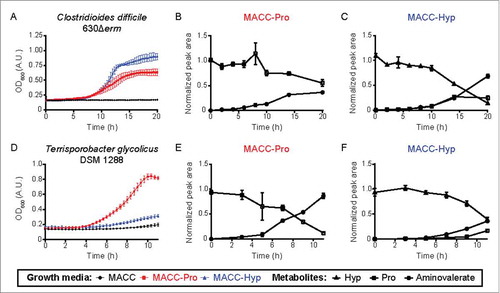
Figure 4. (A) Hyp metabolism by HypD interfaces with microbial amino acid metabolic pathways. Arrows can represent multiple steps and only key metabolites are shown. P5C is a central intermediate in amino acid metabolism. The downstream metabolites α-KG, carbonate, acetyl-CoA, and ammonia can serve as sources of carbon and nitrogen. (B) Sources of Hyp in the human gut from diet or endogenous collagen turnover. Major collagen-derived peptides and Hyp repeats from extensin, a plant cell wall glycoprotein, are shown as examples.
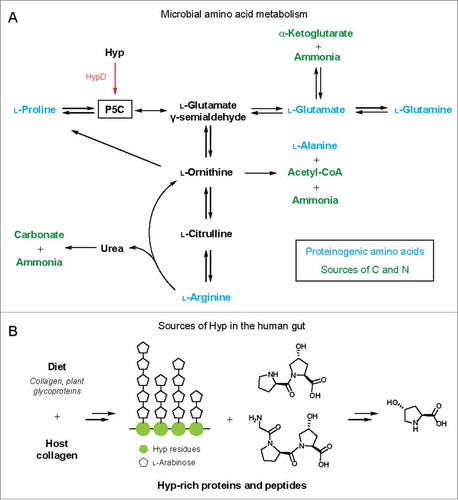
Figure 5. A maximum likelihood tree of 86 full-length HypD amino acid (aa) sequences from the UniProt database. One sequence per species was included. Sequences were aligned with MAFFT using the G-INS-I refinement methodCitation66 and the alignment was trimmed to yield a final length of 783 aa. The tree was constructed in MEGA 767 using the Jones–Taylor–Thorton matrix-based model.Citation68 HypD sequences from species that clade differently from 16S rDNA phylogeny are indicated with an asterisk. Bootstrap values of 70 to 100% are indicated by open circles. Choline trimethylamine-lyase (CutC) and glycerol dehydratase (GD) are included as outgroups.
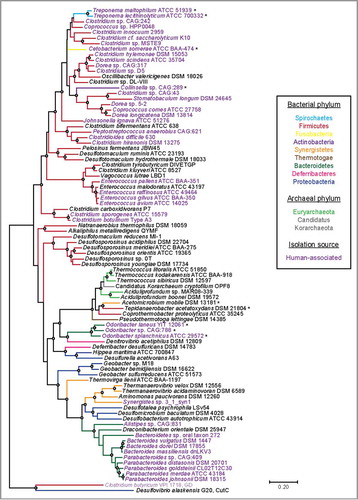
Figure 6. (A) Workflow for enrichment culturing to obtain Hyp-degrading environmental microbes. Hyp-containing supplements were added to certain samples before inoculation into the Hyp enrichment medium. Turbidity and Hyp degradation were observed in all initial cultures before passaging into fresh medium. All cultures were incubated anaerobically at 30ºC in sealed vessels. (B) High Performance Liquid Chromatography-Refractive Index Detector (HPLC-RID) traces for the four enrichments further passaged for sequencing to show Hyp degradation. Samples were derivatized with nitrosonium, generated from potassium nitrite and hydrogen chloride, to obtain N-nitroso derivatives of Pro and Hyp for separation on an Aminex HPX-87P column.Citation69 The previously reported method was modified to include an isocratic elution at 60ºC for 40 minutes.
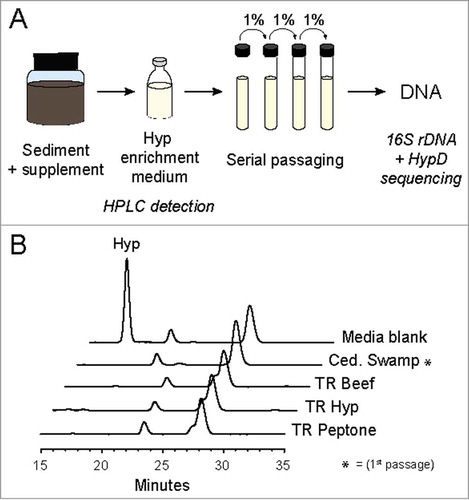
Table 1. All samples were collected at various locations around Woods Hole, MA, USA.
Table 2. Hyp enrichment medium composition.
Table 3. Primers and annealing temperatures used in this study.
Figure 7. 16S rDNA phylogenetic tree of enrichment culture isolates and reference species. 16S rRNA genes were amplified using primers 8F and 1510R () and sequenced from a pCR4-TOPO clone library (Invitrogen). Chimeric, low-quality, and redundant sequences were removed. 28 sequences from enrichment cultures and 12 reference sequences were aligned with MUSCLE.Citation70 The alignment was manually trimmed to yield a final length of 1371 bp and a maximum likelihood tree was constructed in MEGA767 using the Tamura–Nei method of substitution.Citation71 Bootstrap values from 50 to 100% are displayed. The highest similarity score calculated by RDP SeqMatch was used to assign the lowest classification level for each enrichment isolate.Citation72 The full-length sequences obtained for this analysis have been deposited in GenBank (accession: MG367094-MG367121).
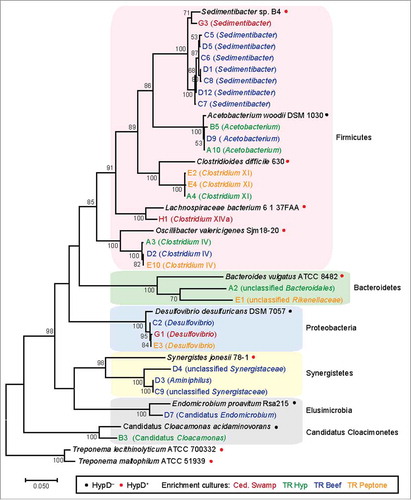
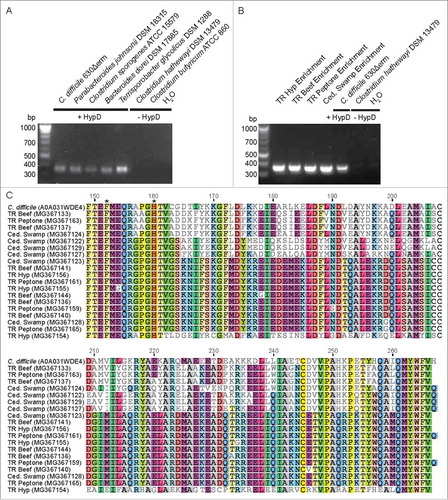
Figure 8. (A) A degenerate primer pair amplified an expected 357 bp product from genomic DNA of HypD-encoding isolates. No amplification product was observed using the genomic DNA of species lacking HypD. (B) Amplification of hypD from 100 ng of DNA from enrichment cultures. Amplicons were sequenced from a clone library generated using TOPO TA cloning kit (Invitrogen). 24 clones from each culture were sequenced by Beckman Coulter Genomics. High quality sequences that are non-redundant within each culture have been deposited on GenBank (accession: MG367122-MG367168). (C) Non-redundant translated amino acid sequences from each enrichment culture were aligned using Clustal OmegaCitation73 along with the biochemically characterized HypD from C. difficile 70-100-2010 (UniProt ID: A0A031WDE4). The two HypD putative active site residues encoded within this amplicon are present in all enrichment sequences and are indicated by asterisks. “Residue numbering is based on HypD sequence from C. difficile 70-100-2010.”