Figures & data
Figure 1. Trehalose metabolism in Clostridium difficile. (A) Canonical metabolism. Under low trehalose conditions the repressor (TreR) remains bound upstream of treA preventing transcription. Under high trehalose concentrations, trehalose-6-phosphate binds to TreR enabling transcription of treA, its subsequent translation and metabolism of trehalose-6-phosphate to glucose and glucose-6-phosphate. (B) In RT027 group strains a conserved mutation in treR, encoding an L172I amino acid substitution near the the effector site, results in increased sensitivity to trehalose-6-phosphate and derepression of treA under low trehalose conditions. (C) RT078 group strains possess an extra 4 gene cassette as well as the canonical treRA operon. The additional trehalose transporter, PtsT, enables efficient transport and metabolism under low trehalose conditions.
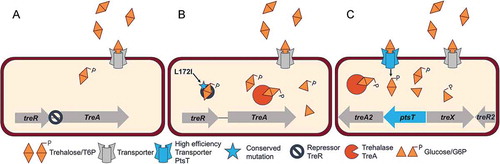
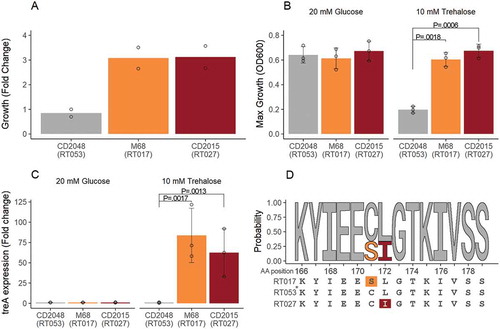
Figure 2. A ribotype 017 strain can utilize low concentrations of trehalose. (A) Biolog 96-well Phenotype MicroArray carbon source plate PM1 was used to examine the metabolic profiles of strains CD2048 (RT053), M68 (RT017) and CD2015 (RT027). Among the carbohydrates tested, strains M68 (RT017) and CD2015 (RT027) displayed a ~ 3 fold increase in growth in the presence of trehalose over no carbohydrate media. CD2048 (RT053) was unable to utilize trehalose. (n = 2) (B) M68 reaches a similar maximum optical density in defined minimal media supplemented with 10 mM trehalose as epidemic ribotype 027. No growth difference is observed in 20 mM glucose. (n = 3). (C) Both M68 (RT017) and CD2015 (RT027) strongly induce treA in the presence of 10 mM trehalose and at a significantly higher level than CD2048 (RT053) . No treA induction is observed in 20 mM glucose.(n = 3). treA expression was normalised to the 16S ribosomal subunit. (D) Multiple sequence alignment of the TreR protein. The consensus sequence for C. difficile near the predicted TreR operator is displayed in large letters while the individual sequences are aligned below. * RT053 sequence is the consensus sequence for all other ribotypes. Figs A-C: All replicates are biological, bars = mean, error = sd.