Figures & data
Figure 1. Luminal presence of supplement-specific probiotics strains in human individuals.
Probiotics strain quantification based on mapping of metagenomic sequences to unique genes, which correspond to the strains found in the supplemented probiotics pill, in the descending colon lumen of healthy individuals treated with an 11-strain probiotic mix (N = 10) or placebo (N = 5). Permissive individuals are those, who were significantly colonized by probiotics in the lower GI mucosa, compared to resistant individuals, in whom no significant colonization of the mucosa was determined. Red, the sample contains the same probiotic strain present in the supplement; Dark gray, the sample contains a different strain of the same species. Strain identification was performed as previously described.Citation111 LAC, Lactobacillus acidophilus; LCA, Lactobacillus casei; LPA, Lactobacillus casei sbsp. paracasei; LPL, Lactobacillus plantarum; LRH, Lactobacillus rhamnosus; BLO, Bifidobacterium longum; BBI, Bifidobacterium bifidum; BBR, Bifidobacterium breve; LLA, Lactococcus lactis; STH, Streptococcus thermophilus.
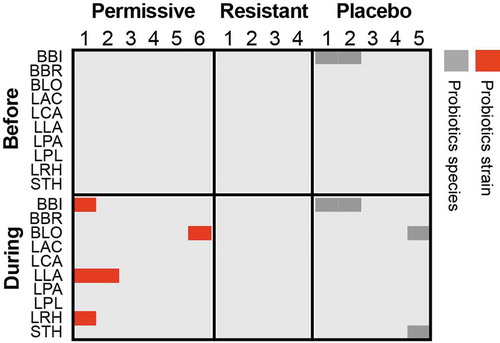
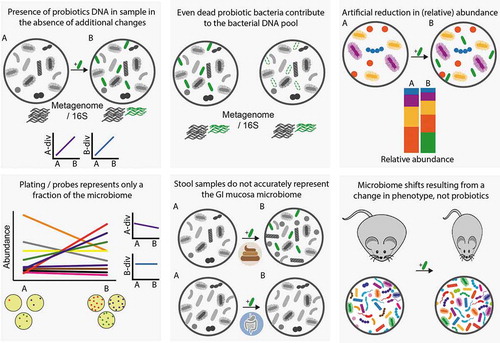
Figure 2. Common biases and challenges observed in studies assessing probiotics effect on microbiome configuration. Supplemented probiotic bacteria are illustrated in green. A-div, alpha diversity; B-div, beta diversity. A, before supplementation; B, during or after supplementation.