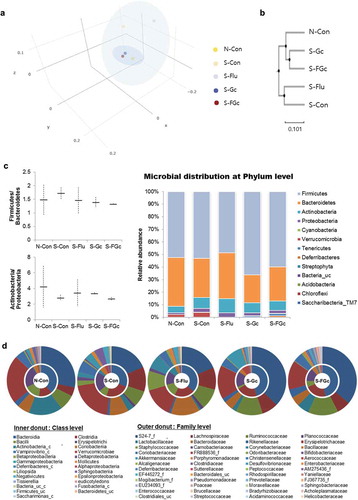
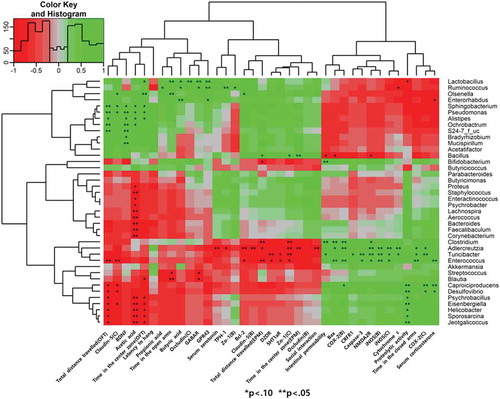
Figures & data
Figure 1. (A) Schematic overview of the in vivo experiment, (B) changes of body weight, food intake, and depressive-like behaviors of mice under UCMS. (C) open-field test, (D) rotarod test, (E) elevated-plus maze test, and (F) social-interaction test. Data are expressed as mean ± SD (n = 8). #Significant difference between N-Con and S-Con (#P < .05, ##P < .005, ###P < .001). *Significant difference with S-Con (*P < .05, **P < .005, ***P < .001).
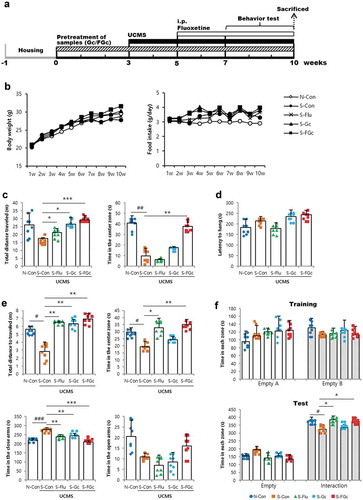
Figure 2. Effects of Gc and FGc on neuroendocrine in mice brain and colon under UCMS. (A) serum corticosterone and serotonin levels (n = 6). (B) Protein expression of NMDA2 R, CRF1 R, 5HT1AR, D2DR, and GABAAR by western blot. Lane 1 = N-Con; lane 2 = S-Con; lane 3 = S-Flu; lane 4 = S-Gc; lane 5 = S-FGc. (C) Protein expression of GPR43 and TPH-1 in mice colon by western blot. Lane 1 = N-Con; lane 2 = S-Con; lane 3 = S-Flu; lane 4 = S-Gc; lane 5 = S-FGc. Data are expressed as mean ± SD (n = 5). #Significant difference between N-Con and S-Con (#P < .05, ##P < .005, ###P < .001). *Significant difference with S-Con (*P < .05, **P < .005, ***P < .001).
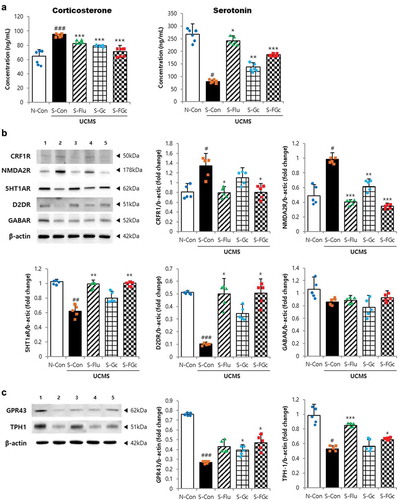
Figure 3. Effects of Gc and FGc on neurodegeneration in mice under UCMS. Expression of proteins related to neurogenesis by western blot. Lane 1 = N-Con; lane 2 = S-Con; lane 3 = S-Flu; lane 4 = S-Gc; lane 5 = S-FGc. Data are expressed as mean ± SD (n = 5). #Significant difference between N-Con and S-Con (#P < .05, ##P < .005, ###P < .001). *Significant difference with S-Con (*P < .05, **P < .005, ***P < .001).
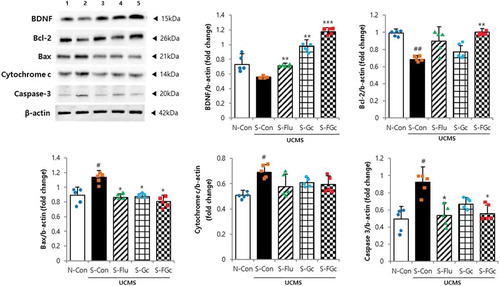
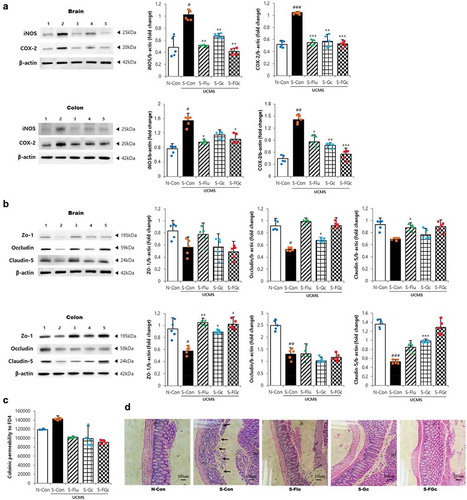
Figure 4. Effects of Gc and FGc on inflammation and barrier function of mice brain and colon under UCMS. (A) Expression of proteins related to inflammation by western blot. Lane 1 = N-Con; lane 2 = S-Con; lane 3 = S-Flu; lane 4 = S-Gc; lane 5 = S-FGc. (B) Expression of proteins related to tight junction by western blot. Lane 1 = N-Con; lane 2 = S-Con; lane 3 = S-Flu; lane 4 = S-Gc; lane 5 = S-FGc. (C) Intestinal epithelial permeability to FD4. (D) Representative H&E stained colon sections. Arrows indicate depleted epithelial cells. Data are expressed as mean ± SD (n = 5). #Significant difference between N-Con and S-Con (#P < .05, ##P < .005, ###P < .001). *Significant difference with S-Con (*P < .05, **P < .005, ***P < .001).