Figures & data
Figure 1. Characteristics of the ETEC infection. ETEC is the major enteric pathogen that account for the diarrhea that occurs in travelers and children in developing countries. ETEC infection is caused by ingestion of contaminated food and water, ETEC through the gastrointestinal tract, and eventually colonization in the small intestine. When ETEC is exposed in the small intestine, it colonizes intestinal epithelial cells via CFs, and ETEC proliferates on the intestinal epithelial after colonization. ETEC produces and delivers heat-labile (LT) and/or heat-stable (ST) enterotoxins to exert toxic effect. Image created with BioRender.
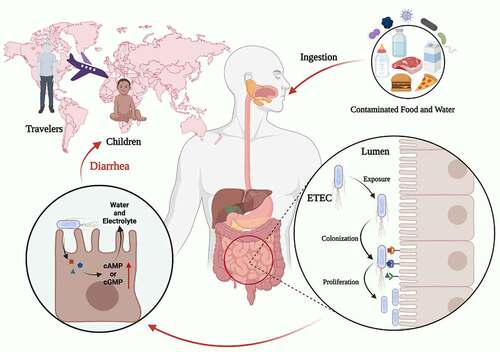
Table 1. Assembly form and morphology of identified colonization factors
Figure 2. The structure of LT, STa, STb, uroguanylin, and guanylin. (a), (b) Three-dimensional structure of the LT (PDB accession no. 1LTB). (c) Three-dimensional structure of the STa (PDB accession no. 1ETN). (d) Three-dimensional structure of the STb (PDB accession no. 1EHS). (e) Three-dimensional structure of the uroguanylin (PDB accession no. 1UYA). (f) Three-dimensional structure of the guanylin (PDB accession no. 1GNA). Image created with BioRender software.
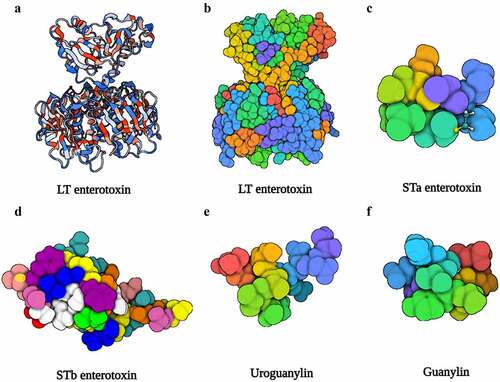
Figure 3. The mechanisms of disease caused by ETEC. Once ETEC established in the small intestinal epithelia via CFs, subsequent efficient enterotoxins delivery activity begins. The ST and LT of ETEC activate adenylyl and guanylate cyclase lead to high level of cAMP and cGMP, which stimulates water and electrolytes secretion in the intestinal lumen.
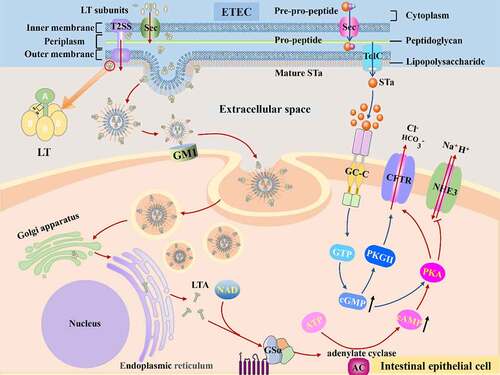
Table 2. Characteristic of LT produced by ETEC
Table 3. Characteristic of ST produced by ETEC
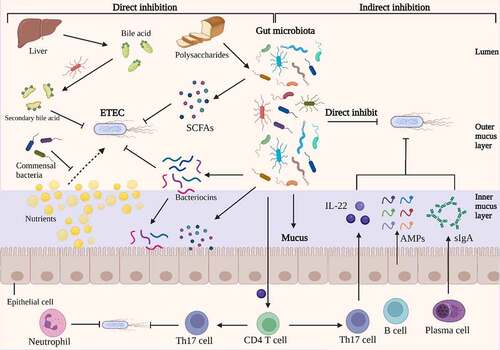
Figure 4. Direct and indirect inhibition mediated by gut microbiota against ETEC infections. On the left, an illustration depicts the direct inhibition against ETEC mediated by gut bacteria. Gut bacteria directly impede ETEC colonization and proliferation. Certain antibacterial compounds, such as bacteriocins, SCFAs, and secondary bile acid, generated by the gut microbiota have been shown to directly inhibit ETEC. Additionally, gut microbiota can compete with ETEC for nutrients, which could limit the growth of ETEC. On the right, indirect methods of competition between gut microbiota and ETEC are depicted. The antimicrobial molecules produced by gut microbiota, such as SCFAs and bacteriocins, which could release into inner mucus layer and stimulate the barrier function. The commensal microbiota induces the differentiation of CD4 T cells into Th17 cells, which contribute to colonization resistance against ETEC by the release of cytokines such as IL-22. Under the stimulation of gut microbiota, intestinal epithelia secrete inflammatory factors, AMPs and sIgA into the mucus, which inhibits the colonization of ETEC. Image created with BioRender.