Figures & data
Figure 1. Structures of the repeat units in various GAGs. (A) HA is an unsulfated polymer with [−4)-β-GlcUA-(1-3)-β-GlcNAc-(1-] as the repeating unit. (B) CS is the polymer of sulfated [−4)-β-GlcUA-(1-3)-β-GalNAc-(1-]. Depending on the sulfation pattern, CS can be classified into different subtypes (CSA, CSC, CSD, and CSE). (C) DS composes of the repeating dimeric unit [−4)-α-IdoUA-(1-3)-β-GalNAc-(1-]. L-IdoUA is an epimer of D-GlcUA and the GalNAc residue is sulfated at different positions similar to CS. (D) The major disaccharide unit of heparin is [−4)-α-IdoUA-(1-4)-α-GlcN-(1-]. Iduronic acid is sulfated at O2 and the GlcN unit is N- and O6-sulfated. (E) The major disaccharide unit of HS is [−4)-α-GlcUA-(1-4)-β-GlcNAc-(1-]. The GlcNAc residue is generally sulfated at N2 with or without O6 sulfation. (F) In the typical KS, disaccharide subunits are composed of Gal and GlcNAc-6-sulfate. The Gal residues may or may not have O6 sulfation.
![Figure 1. Structures of the repeat units in various GAGs. (A) HA is an unsulfated polymer with [−4)-β-GlcUA-(1-3)-β-GlcNAc-(1-] as the repeating unit. (B) CS is the polymer of sulfated [−4)-β-GlcUA-(1-3)-β-GalNAc-(1-]. Depending on the sulfation pattern, CS can be classified into different subtypes (CSA, CSC, CSD, and CSE). (C) DS composes of the repeating dimeric unit [−4)-α-IdoUA-(1-3)-β-GalNAc-(1-]. L-IdoUA is an epimer of D-GlcUA and the GalNAc residue is sulfated at different positions similar to CS. (D) The major disaccharide unit of heparin is [−4)-α-IdoUA-(1-4)-α-GlcN-(1-]. Iduronic acid is sulfated at O2 and the GlcN unit is N- and O6-sulfated. (E) The major disaccharide unit of HS is [−4)-α-GlcUA-(1-4)-β-GlcNAc-(1-]. The GlcNAc residue is generally sulfated at N2 with or without O6 sulfation. (F) In the typical KS, disaccharide subunits are composed of Gal and GlcNAc-6-sulfate. The Gal residues may or may not have O6 sulfation.](/cms/asset/3c9e401f-f4d0-41e2-bddb-cce0349e43e0/kgmi_a_2068367_f0001_oc.jpg)
Figure 2. The scheme of chondroitin sulfate degradation by Bacteroides thetaiotaomicron VPI-5482. Turnover of gut epithelium and the diet are the two sources of CS in the gut. The surface-localized BT3328-PL29 dissimilates extracellular CS to produce oligomers, which are bound by SGBPs: BT3329, BT3330, and BT3332 (SusD-like), and internalized by BT3331-SusC-like transporter. These oligomers are processed by three periplasmic PLs (BT3324-PL8, BT3350-PL8, and BT4410-PL33) and BT3349-4-O-sulfatase (S1_27). The disaccharide products of these CS lyases are desulfated by BT1596 (S1_9) at the 2-O position. The GH88 Δ-4,5-unsaturated β-glucuronyl hydrolase (BT3348) processes these disaccharides to hexuronic acid and GalNAc or GalNAc6S monomers. BT3333-sulfatase (S1_15) removes the 6-O-Sulfate group from the GalNAc6S residues if required. All the unsulfated monomers are imported via the inner membrane into the cytoplasm for utilization. The HTCS protein BT3334 is activated by the BT1596 (S1_9) product.
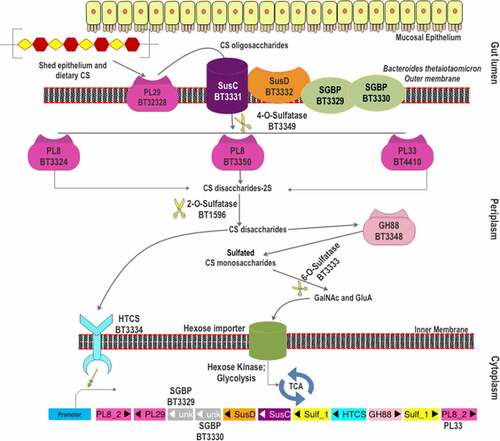
Figure 3. The gene organization of the CS-PUL in various gut Bacteroides. The CS-PUL in B. thetaiotaomicron spans from bt3324-bt3350, and a distant bt4410 gene was also involved in CS degradation. PLs (purple), SusD and SGBP (orange), SusC (plum), GH88 (pink), sulfatase (yellow), and HTCS (cyan) encoded in PULs are indicated. Genome accession numbers are presented at the top of each Bacteroides species, and gene numbers of various genes are provided below each gene.

Figure 4. The scheme of heparin and HS degradation by Bacteroides thetaiotaomicron VPI-5482. Heparin/HS shed from epithelial cell line and acquired from the diet enter the gut lumen. A surface HS lyase (BT4662-PL12) depolymerizes these glycans into oligosaccharides, which are then internalized by BT4660-SusC via binding to BT4659-SusD and BT4661-SGBP. In the periplasm, three more heparin/HS lyases (BT4652-PL15, BT4657-PL12, and BT5475-PL13) cleave these oligomers to produce disaccharides. The disaccharides are desulfated at the 2-O position by BT1596 (S1_9). The product of sulfatase BT1596 is the substrate of the GH88 Δ-4,5-unsaturated β-glucuronyl hydrolase BT4658 and the activator ligand of HTCS protein. The monosaccharides liberated by GH88 are finally desulfated by BT4656-6-O-sulfatase (S1_11) and a yet unknown 3-O-sulfatase. The unsulfated monomers are imported into the cytoplasm for further metabolism. Hep: heparin.
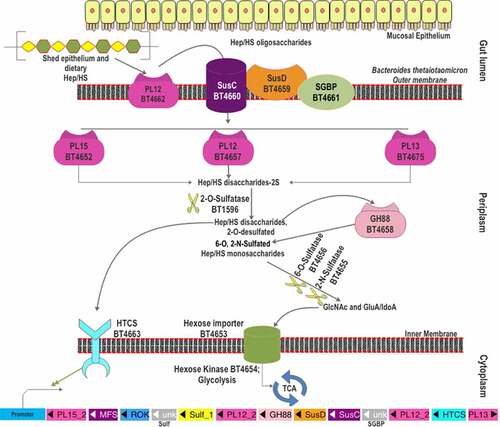
Figure 5. The gene organization of the Heparin/HS-PUL in various gut Bacteroides. The HS-PUL in B. thetaiotaomicron VPI-5482 spans from bt4652-bt4675. PLs (purple), SusD and SGBP (Orange), SusC and MFS (plum), GH88 (pink), sulfatase (yellow), HTCS (cyan), and ROK (blue) encoded in PULs are indicated. Genome accession numbers are presented at the top of each Bacteroides species and gene numbers of various genes are provided below each gene.

Figure 6. Putative GAG-utilization genomic loci in non-Bacteroidetes gut bacteria. Except for Roseburia hominis A2-183, other species have been shown to degrade GAGs. The heparin gpPUL in Roseburia hominis A2-183 is predicted by Sheridan et al., 2016. The gpPUL of S. pneumoniae R6 has been characterized by Marion et al., 2012. Other PULs are not reported in the literature and are identified according to the definition of PUL/gpPUL by searching their genomic assemblies. Genome accession numbers are presented at the top of each species. PL8: putative chondroitinase/hyaluronidase; KdgA: 2-keto-3-deoxygluconate aldolase: KdgK: 2-keto-3-deoxygluconate kinase; DhuI: 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase; DhuD: 2-keto-3-deoxy-D-gluconate dehydrogenase; EIIA-EIID: Enzyme complex II subunit A to D of PTS system; PL12: putative heparinase; GalR: galactose operon repressor type transcription regulator; LaciR: lactose operon repressor type transcription regulator; GH27, GH43: glycoside hydrolase family 27 and 43 proteins; TCS: two-component response regulatory system; HK sensor: sensor protein of the Histidine kinase sensor system; ABC: ABC transporter proteins; Spi: sugar-phosphate isomerase; SGBP: surface glycan-binding protein; FBA: fructose biphosphate aldolase class II; Rpe: ribulose phosphate isomerase; BgiG: transcription anti-terminator; GntR: gluconate-operon repressor type transcription regulator; ΔGH: unsaturated glucuronyl hydrolase; Eda: Entner-Doudoroff aldolase: IclR: glyoxylate bypass repressor type transcription regulator; kduI: 5-dehydroxy-4deoxy-D-glucuronate Isomerase; RpiR: HTH-type transcriptional regulator; Sul: sulfatase; kduD: 5-dehydroxy-4deoxy-D-glucuronate dehydrogenase.

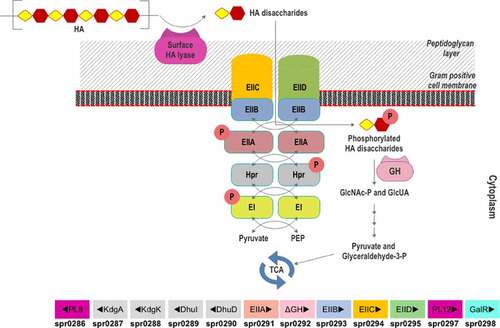
Figure 7. Scheme of HA utilization by Streptococcus pneumoniae R6. Extracellular HA is acquired and fragmented by a surface hyaluronidase to unsaturated HA disaccharides. Internalization of HAΔdi occurs via the phosphorylation-dephosphorylation cascade of PTS subunits (EI and EIIA-EIID), HPr protein, pyruvate, and phosphoenolpyruvate (PEP). The GlcNAc-phosphorylated HAΔdi is decomposed into monosaccharides (GlcA and GlcNAc-P) in the cytoplasm by an unsaturated glycoside hydrolase (GH). The monosaccharides are funneled through various enzymatic reactions to the energy production pathway. The HA-gpPUL with the gene numbers of the bacteria is shown at the bottom. PL8: hyaluronidase; KdgA: 2-keto-3-deoxygluconate aldolase: KdgK: 2-keto-3-deoxygluconate kinase; DhuI: 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase; DhuD: 2-keto-3-deoxy-D-gluconate dehydrogenase; EIIA-EIID: Enzyme complex II subunit A to D of PTS system; PL12: putative heparinase; GalR: galactose operon repressor type transcription regulator; EI: Enzyme complex I of PTS system; Hpr: Hpr protein of PTS system.