Figures & data
Figure 1. Change of alpha and beta diversity between outcomes. (a) The observed richness and Shannon diversity for responders, non-responders, and donor samples. Alphabet letters are the Tukey’s post -hoc test; sharing same letters indicate no significant difference at a level of 0.05. (b) The change of observed richness and Shannon diversity within patients and averaged by outcomes. The error bars refer to the 95% confidence interval of the mean estimated with paired Welch’s t-test. Error bars not overlapping the zero value indicate significant changes compared to pre-FMT (W0). (c) The beta diversity of samples from response and non-response patients assessed with Bray-Curtis distance and visualized with nonmetric multidimensional scaling (NMDS) ordination. Shapes refer to time points or donor. Colors refer to outcomes or donor. Samples are shown with small shapes referring to different time points. The big shapes represent the centroid of sample groups in each condition and connected with lines in the order of time point. (d) Bray-Curtis distance between patient samples and corresponding donor samples. Each dot refers to one distance between a given patient and the processed feces coming from the corresponding donor. The crossbars represent mean value distances. P values are obtained with the Welch’s t-test. For (a) and (c), the taxa abundances in fecal samples coming from the same donor (in total six donors provided 24 samples) were averaged so that each donor is represented by a single community.
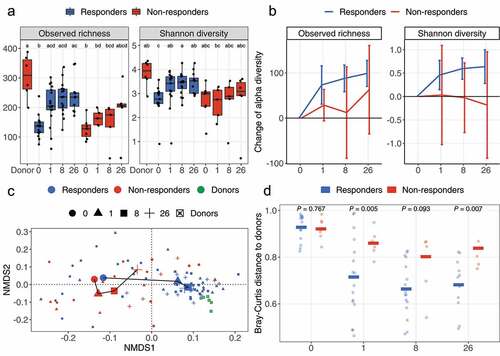
Figure 2. Tree plot showing taxa with different relative abundances between outcomes. The largest node in the center is the kingdom bacteria. Along the tree branch outward, the next node is the phylum level, then followed by class, order, family, and genus. The size of the node is proportional to the mean relative abundance in the corresponding phylogenetic level. A node is labeled when it has significantly different relative abundances (Wilcoxon rank-sum test) between outcomes. Blue refers to higher abundances in responders, red refers to lower abundances in responders. The three tree plots represent data from Week 0, 1, and 8.
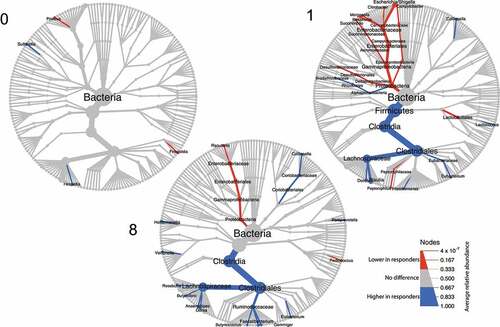
Figure 3. Engraftment of donor microbes and improvement of dysbiosis score. (a) Engraftment of donor microbes was estimated with the SourceTracker program using each patient as the “sink” and the corresponding donor as the “source”. Lines crossing time points represent the mean similarity in the microbial communities between donors and patients. The shaded area indicates standard error of the mean. P values were obtained by Welch’s t-test. (b) The top 10 most engrafted families. ASVs engrafted in patients were identified with SourceTracker and their relative abundances were summed at the family level. The top 10 most abundant families are shown and the remaining are merged as “Others”. (c) Dysbiotic status of patients as estimated with the CLOUD test. Patients (test sample) were tested against the donors (reference) using CLOUD, and the derived statistic from the test was used as the dysbiosis score, where smaller scores correspond to healthier bacterial communities. The differences in dysbiosis scores between outcomes were compared with Wilcoxon rank-sum test.
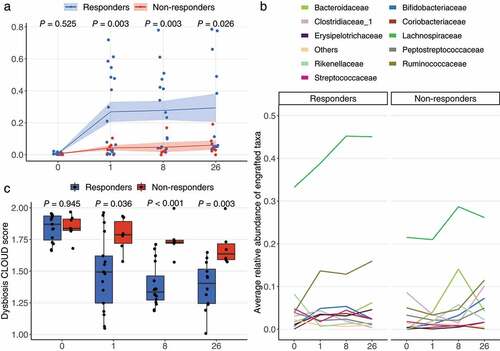
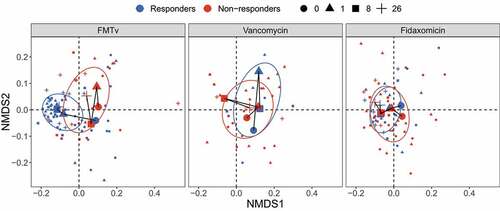
Figure 4. Beta diversity of fecal microbiota for FMTv, vancomycin, and fidaxomicin treatments. Beta diversity is assessed based on the Bray-Curtis distance and visualized by NMDS ordination. Colors of dots and ellipses (50% confidence regions for clusters) refer to different outcomes. Samples are shown with small shapes referring to different time points. Big shapes represent the centroids of sample groups in each condition and are connected with lines according to sample time.
Supplemental Material
Download PDF (1.5 MB)Data availability statement
The raw sequence data of 16S rRNA gene sequencing reported in this paper are available from the National Center for Biotechnology Information (NCBI) with the Sequence Read Archive (SRA) bioproject number PRJNA797470. All other data and the code used to reanalyze the data reported in this paper is available from the corresponding author upon request. ncbi.nlm.nih.gov/sra/?term=PRJNA797470
