Figures & data
Figure 1: Distinct fecal microbiota profiles of HBV-CLD patients and HCs. Flow chart of the study design (a); β diversity (based on Bray-Curtis distances) evaluated by PCoA analysis (b); PLS-DA analysis (c). HBV-CLD, chronic hepatitis B virus infection-associated liver diseases; HC, healthy control; HCC, hepatocellular carcinoma; LC-MS, liquid chromatography–mass spectrometry; T2DM, diabetes mellitus type 2.
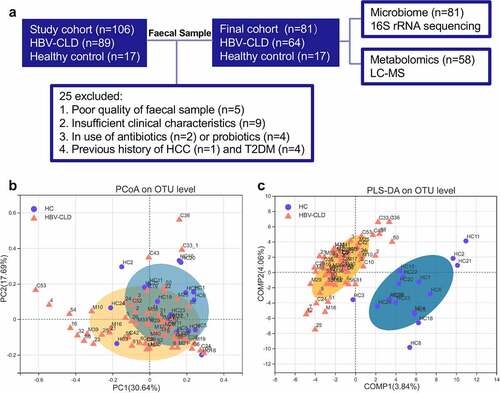
Figure 2: Gut microbiota profile in HBV-CLD patients. The abundance of genera was log-transformed with 0 values assigned with 1e-05. Box plots indicate median (middle line), 25th, 75th percentile (box) and maximum and minimum values (whisker). *pfdr<0.05, **pfdr<0.01, ***pfdr<0.001, ****pfdr<0.0001. HBV-CLD, chronic hepatitis B virus infection-associated liver diseases; HC, healthy control.
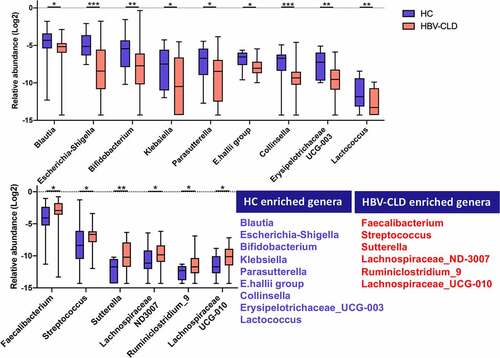
Figure 3: Distinct gut microbiota profile regarding disease progression and antiviral treatment. Genera correlated with disease progression (a); antiviral treatment (b); Venn diagram outlined the genera associated with disease progression and antiviral treatment (c). The abundance of genera was log-transformed with 0 values assigned with 1e-05. Box plots indicate median (middle line), 25th, 75th percentile (box) and maximum and minimum values (whisker). *pfdr<0.05, **pfdr<0.01, ***pfdr<0.001, ****pfdr<0.0001. CHB, chronic hepatitis B (treatment naive); Crrh, cirrhosis; ETV, entecavir; HC, healthy control; NC, non-cirrhosis.
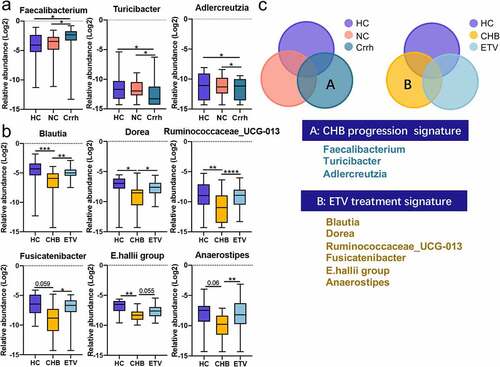
Figure 4: Distinct fecal metabolome signature of HBV-CLD patients and HCs. Representative metabolites that were significantly changed in HBV-CLD (a); correlated with disease progression (b); antiviral treatment (c); Venn diagram outlined the metabolites associated with disease progression and antiviral treatment (d). The abundance of metabolites was log-transformed with 0 values assigned with 1e-05. Box plots indicate median (middle line), 25th, 75th percentile (box) and maximum and minimum values (whisker). *pfdr<0.05, **pfdr<0.01, ***pfdr<0.001, ****pfdr<0.0001. CHB, chronic hepatitis B (treatment naive); Crrh, cirrhosis; ETV, entecavir; HBV-CLD, hepatitis B virus related chronic liver diseases; HC, healthy control; NC, non-cirrhosis.
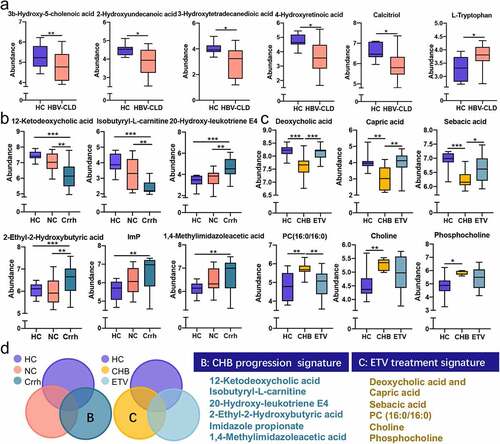
Figure 5: The host-microbiota-metabolite interplay in HBV-CLD patients. The heatmap depicts correlation (partial spearman analysis) of disease-related taxa and metabolites (a); disease-related metabolites and clinical indexes (b); disease-related metabolites and clinical indexes (c). *pfdr<0.05, **pfdr<0.01, ***pfdr<0.001. ALB, albumin; ALT, alanine amino transferase; AST, aspartate aminotransferase; GGT, γ-glutamyl transpeptidase; TB, total bilirubin; DB, direct bilirubin; PT, prolonged prothrombin time.
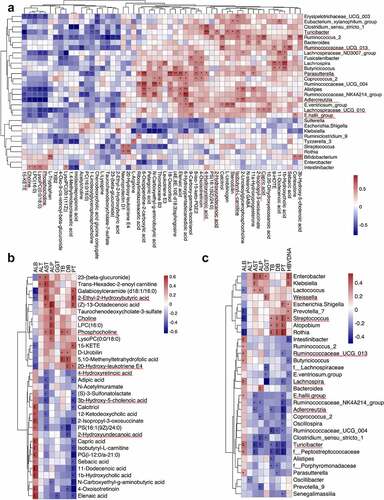
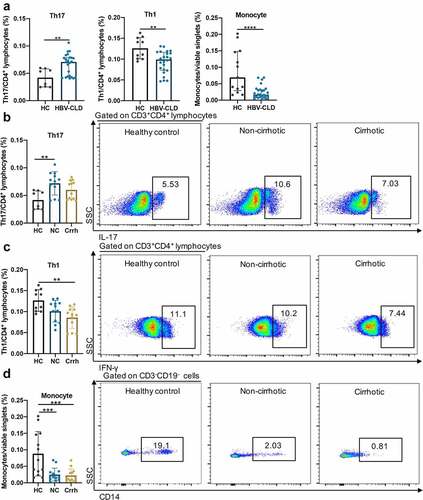
Figure 6: Exposure of PBMCs to patient-derived BEs resulted in altered T cell subtype counts. Th17 (CD3+CD4+IL-17A+), Th1 (CD3+CD4+IFN-γ+), monocytes (CD3−CD19−CD14+) in response to BE exposure from HCs and HBV-CLD patients (a). Th17 (b), Th1 (c) and monocytes (d) in response to BE exposure from HCs, non-cirrhotic (NC) and cirrhotic (Crrh) patients. Data are shown as % of viable CD3+CD4+ lymphocytes for Th17 and Th1 cells, and % of viable singlets for monocytes. Box plots indicate individual values for each sample and median with interquartile range within groups. *p < .05, **p < .01, ***p < .001, ****p < .0001. BE, bacterial extracts; Crrh, cirrhosis; HBV-CLD, hepatitis B virus related chronic liver diseases; HC, healthy control; NC, non-cirrhosis; Th1, T helper 1; Th17, T helper 17.