Figures & data
Figure 1. The T6SS2 secretome of V. parahaemolyticus strain BB22OP. (a) A volcano plot summarizing the comparative proteomics of proteins identified in the medium of wild type (WT) and T6SS2− (∆hcp2) V. parahaemolyticus BB22OP strains using label-free quantification. The average difference in signal intensities between the WT strain and the ∆hcp2 strain is plotted against the -Log10 of Student’s t-test P values (n = 3 biological replicates). Proteins that were significantly more abundant in the secretome of the WT strain (difference in the average LFQ intensities > 2; P value < .03; with a minimum of 5 Razor unique peptides) are denoted in red and annotated. (b) Schematic representation of genome neighborhoods for non-structural T6SS2-secreted proteins identified in (a). Predicted secreted effectors are denoted in red; predicted neighboring immunity genes are denoted in cyan. Arrows indicate the direction of transcription, and the names of encoded proteins or domains are denoted below. The RefSeq GenBank accession number and the locus tag range are provided.
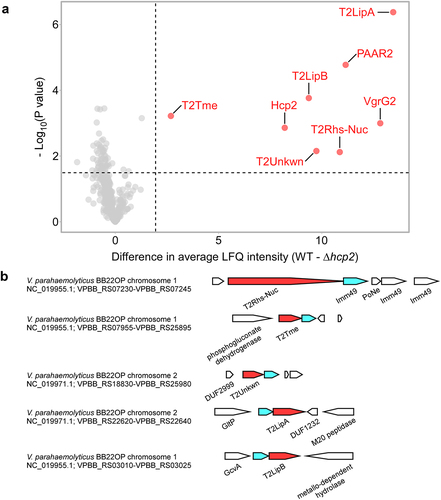
Table 1. Predicted effectors secreted by V. parahaemolyticus T6SS2.
Figure 2. The T6SS2 secretome of V. parahaemolyticus strain RIMD 2210633. (a) Expression (cells) and secretion (media) of Hcp2 from the V. parahaemolyticus RIMD 2210633 strain containing an empty plasmid (pTfoX -) or a plasmid for the arabinose-inducible expression of a C-terminal Myc-His tagged TfoX (pTfoX +). Samples were grown in LB media supplemented with kanamycin to maintain the plasmids, and in the presence (+) or absence (-) of phenamil at 30°C. Loading control (LC) is shown for total protein lysates. (b) Viability counts (colony forming units [CFU]) of V. natriegens prey strain before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus RIMD 2210633 attacker strain carrying an empty plasmid (pEmpty) or a plasmid for the arabinose-inducible expression of TfoX (pTfoX) on LB agar plates supplemented with 0.1% (wt/vol) L-arabinose to induce expression from plasmids. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test. Data are shown as the mean ± SD; n = 3. (c) A volcano plot summarizing the comparative proteomics of proteins identified in the medium of the T6SS2+ (∆hcp1) and T6SS2− (∆hcp1∆hcp2) V. parahaemolyticus RIMD 2210633 strains, expressing TfoX from a plasmid, using label-free quantification. The average difference in the signal intensities between the T6SS2+ strain and the T6SS2− strain is plotted against the -Log10 of Student’s t-test P values (n = 3 biological replicates). Proteins that were significantly more abundant in the secretome of the T6SS2+ strain (difference in the average LFQ intensities > 2; P value < .03; with a minimum of 5 Razor unique peptides) are denoted in red and annotated. (d) Schematic representation of genome neighborhoods for non-structural T6SS2-secreted proteins identified in (c). Predicted secreted effectors are denoted in red; predicted neighboring immunity genes are denoted in cyan; vp2395, which is not predicted to be an effector, is denoted in orange. Arrows indicate the direction of transcription, and the names of encoded proteins or domains are denoted below. The RefSeq GenBank accession number and the locus tag range are provided.
![Figure 2. The T6SS2 secretome of V. parahaemolyticus strain RIMD 2210633. (a) Expression (cells) and secretion (media) of Hcp2 from the V. parahaemolyticus RIMD 2210633 strain containing an empty plasmid (pTfoX -) or a plasmid for the arabinose-inducible expression of a C-terminal Myc-His tagged TfoX (pTfoX +). Samples were grown in LB media supplemented with kanamycin to maintain the plasmids, and in the presence (+) or absence (-) of phenamil at 30°C. Loading control (LC) is shown for total protein lysates. (b) Viability counts (colony forming units [CFU]) of V. natriegens prey strain before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus RIMD 2210633 attacker strain carrying an empty plasmid (pEmpty) or a plasmid for the arabinose-inducible expression of TfoX (pTfoX) on LB agar plates supplemented with 0.1% (wt/vol) L-arabinose to induce expression from plasmids. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test. Data are shown as the mean ± SD; n = 3. (c) A volcano plot summarizing the comparative proteomics of proteins identified in the medium of the T6SS2+ (∆hcp1) and T6SS2− (∆hcp1∆hcp2) V. parahaemolyticus RIMD 2210633 strains, expressing TfoX from a plasmid, using label-free quantification. The average difference in the signal intensities between the T6SS2+ strain and the T6SS2− strain is plotted against the -Log10 of Student’s t-test P values (n = 3 biological replicates). Proteins that were significantly more abundant in the secretome of the T6SS2+ strain (difference in the average LFQ intensities > 2; P value < .03; with a minimum of 5 Razor unique peptides) are denoted in red and annotated. (d) Schematic representation of genome neighborhoods for non-structural T6SS2-secreted proteins identified in (c). Predicted secreted effectors are denoted in red; predicted neighboring immunity genes are denoted in cyan; vp2395, which is not predicted to be an effector, is denoted in orange. Arrows indicate the direction of transcription, and the names of encoded proteins or domains are denoted below. The RefSeq GenBank accession number and the locus tag range are provided.](/cms/asset/05b162e2-6328-4261-9aaf-a4f307244aa2/kgmi_a_2178795_f0002_oc.jpg)
Figure 3. Validation of V. parahaemolyticus strain BB22OP T6SS2 effector and immunity pairs. Viability counts (CFU) of the indicated BB22OP derivative prey strains containing a deletion of the predicted effectors T2Rhs-NucBB22 (a), T2TmeBB22 (b), T2UnkwnBB22 (c), T2LipABB22 (d), and T2LipBBB22 (e), and their neighboring predicted immunity gene (-i) before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus BB22OP attacker strains. Prey strains contain either an empty plasmid (Empty) or a plasmid for the arabinose-inducible expression of the predicted immunity protein that was deleted (Immunity). Competitions were performed on LB agar plates supplemented with L-arabinose (0.1% [wt/vol] in A, B, C, and E; 0.01% [wt/vol] in D) to induce protein expression. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test; ns, no significant difference (P > .05). Data are shown as the mean ± SD; n = 3.
![Figure 3. Validation of V. parahaemolyticus strain BB22OP T6SS2 effector and immunity pairs. Viability counts (CFU) of the indicated BB22OP derivative prey strains containing a deletion of the predicted effectors T2Rhs-NucBB22 (a), T2TmeBB22 (b), T2UnkwnBB22 (c), T2LipABB22 (d), and T2LipBBB22 (e), and their neighboring predicted immunity gene (-i) before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus BB22OP attacker strains. Prey strains contain either an empty plasmid (Empty) or a plasmid for the arabinose-inducible expression of the predicted immunity protein that was deleted (Immunity). Competitions were performed on LB agar plates supplemented with L-arabinose (0.1% [wt/vol] in A, B, C, and E; 0.01% [wt/vol] in D) to induce protein expression. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test; ns, no significant difference (P > .05). Data are shown as the mean ± SD; n = 3.](/cms/asset/d8eb47bc-f45a-4fda-a152-7802f7a493cf/kgmi_a_2178795_f0003_oc.jpg)
Figure 4. Validation of V. parahaemolyticus strain RIMD 2210633 T6SS2 effector and immunity pairs. Viability counts (CFU) of the indicated RIMD 2210633 derivative prey strains containing a deletion of the predicted effectors T2Rhs-NucRIMD (a), T2HydroRIMD (b), and T2LipBRIMD (c), and their neighboring predicted immunity gene (-i) before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus RIMD 2210633 attacker strains. Prey strains contain either an empty plasmid (Empty) or a plasmid for the arabinose-inducible expression of the predicted immunity protein that was deleted (Immunity). Competitions were performed on LB agar plates supplemented with 0.1% [wt/vol] L-arabinose to induce protein expression. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test; ns, no significant difference (P > .05). Data are shown as the mean ± SD; n = 3.
![Figure 4. Validation of V. parahaemolyticus strain RIMD 2210633 T6SS2 effector and immunity pairs. Viability counts (CFU) of the indicated RIMD 2210633 derivative prey strains containing a deletion of the predicted effectors T2Rhs-NucRIMD (a), T2HydroRIMD (b), and T2LipBRIMD (c), and their neighboring predicted immunity gene (-i) before (0 h) and after (4 h) co-incubation with the indicated V. parahaemolyticus RIMD 2210633 attacker strains. Prey strains contain either an empty plasmid (Empty) or a plasmid for the arabinose-inducible expression of the predicted immunity protein that was deleted (Immunity). Competitions were performed on LB agar plates supplemented with 0.1% [wt/vol] L-arabinose to induce protein expression. The statistical significance between samples at the 4 h time point was calculated using an unpaired, two-tailed Student’s t-test; ns, no significant difference (P > .05). Data are shown as the mean ± SD; n = 3.](/cms/asset/6bb9a60d-9edb-44ed-b979-7d330886330b/kgmi_a_2178795_f0004_oc.jpg)
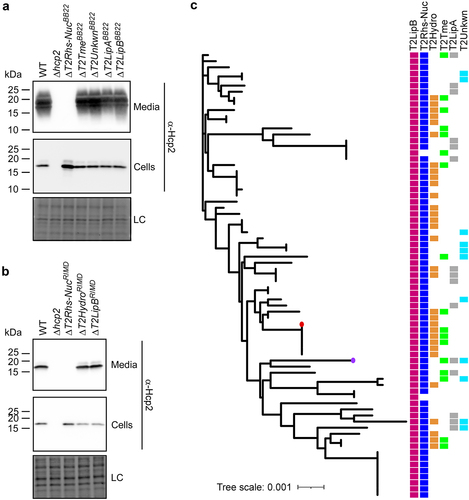
Figure 5. T2Rhs-Nuc is ubiquitous in V. parahaemolyticus genomes and is required for T6SS2 activity. (a-b) Expression (cells) and secretion (media) of Hcp2 from V. parahaemolyticus BB22OP (a) and RIMD 2210633 (b) wild-type (WT) strains or their indicated derivatives containing a deletion in a gene encoding a secreted T6SS2 effector. Samples were grown in LB media at 30°C. Loading control (LC) is shown for total protein lysates. (c) Distribution of T6SS2-secreted effectors in complete V. parahaemolyticus genomes. The phylogenetic tree was based on DNA sequences of rpoB coding for DNA-directed RNA polymerase subunit beta. The evolutionary history was inferred using the neighbor-joining method. V. parahaemolyticus strains BB22OP and RIMD 2210633 are denoted by a circle (purple and red, respectively).
Supplemental Material
Download Zip (1.2 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material. The mass spectrometry raw data files were deposited in ProteomeXchange under the accession numbers indicated in the Materials and Methods section. The data can be accessed via the following links: For strain BB22OP http://www.ebi.ac.uk/pride/archive/projects/PXD037864; for strain RIMD 2210633 http://www.ebi.ac.uk/pride/archive/projects/PXD037980.
